[English] 日本語
 Yorodumi
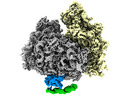
Yorodumi- EMDB-10609: Cryo-EM structure of the human Ebp1-ribosome complex (Ebp1-ES27L ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10609 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human Ebp1-ribosome complex (Ebp1-ES27L segment) | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.7 Å | |||||||||
 Authors Authors | Aleksic M / Wild K / Sinning I / Pfeffer S | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: MetAP-like Ebp1 occupies the human ribosomal tunnel exit and recruits flexible rRNA expansion segments. Authors: Klemens Wild / Milan Aleksić / Karine Lapouge / Keven D Juaire / Dirk Flemming / Stefan Pfeffer / Irmgard Sinning /  Abstract: Human Ebp1 is a member of the proliferation-associated 2G4 (PA2G4) family and plays an important role in cancer regulation. Ebp1 shares the methionine aminopeptidase (MetAP) fold and binds to mature ...Human Ebp1 is a member of the proliferation-associated 2G4 (PA2G4) family and plays an important role in cancer regulation. Ebp1 shares the methionine aminopeptidase (MetAP) fold and binds to mature 80S ribosomes for translational control. Here, we present a cryo-EM single particle analysis reconstruction of Ebp1 bound to non-translating human 80S ribosomes at a resolution range from 3.3 to ~8 Å. Ebp1 blocks the tunnel exit with major interactions to the general uL23/uL29 docking site for nascent chain-associated factors complemented by eukaryote-specific eL19 and rRNA helix H59. H59 is defined as dynamic adaptor undergoing significant remodeling upon Ebp1 binding. Ebp1 recruits rRNA expansion segment ES27L to the tunnel exit via specific interactions with rRNA consensus sequences. The Ebp1-ribosome complex serves as a template for MetAP binding and provides insights into the structural principles for spatial coordination of co-translational events and molecular triage at the ribosomal tunnel exit. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10609.map.gz emd_10609.map.gz | 2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10609-v30.xml emd-10609-v30.xml emd-10609.xml emd-10609.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10609.png emd_10609.png | 203.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10609 http://ftp.pdbj.org/pub/emdb/structures/EMD-10609 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10609 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10609 | HTTPS FTP |
-Validation report
| Summary document |  emd_10609_validation.pdf.gz emd_10609_validation.pdf.gz | 198.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10609_full_validation.pdf.gz emd_10609_full_validation.pdf.gz | 197.9 KB | Display | |
| Data in XML |  emd_10609_validation.xml.gz emd_10609_validation.xml.gz | 6.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10609 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10609 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10609 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10609 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10609.map.gz / Format: CCP4 / Size: 181 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10609.map.gz / Format: CCP4 / Size: 181 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5218 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human Ebp1-ribosome complex
| Entire | Name: Human Ebp1-ribosome complex |
|---|---|
| Components |
|
-Supramolecule #1: Human Ebp1-ribosome complex
| Supramolecule | Name: Human Ebp1-ribosome complex / type: complex / ID: 1 / Parent: 0 Details: Puromycin-treated ribosomes from HeLa cells in complex with recombinantly expressed Ebp1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 20 mM HEPES KOH pH 7.5 5 mM Mg(OAc)2 175 mM KOAc 1 mM Tris(2-carboxyethyl)phosphin |
|---|---|
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
| Details | 100 nM ribosome; 800 nM Ebp1 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 3370 / Average electron dose: 38.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)