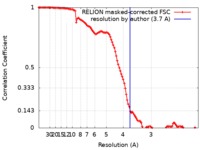
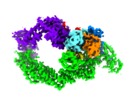
登録情報 データベース : EMDB / ID : EMD-10585タイトル Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2 複合体 : Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2複合体 : S-phase kinase-associated protein 1タンパク質・ペプチド : E3 ubiquitin-protein ligase RBX1複合体 : Cullin-1, E3 ubiquitin-protein ligase RBX1タンパク質・ペプチド : S-phase kinase-associated protein 1タンパク質・ペプチド : NEDD8複合体 : F-box/WD repeat-containing protein 1A, NEDD8, S-phase kinase-associated protein 1, Ubiquitin-conjugating enzyme E2 D2タンパク質・ペプチド : F-box/WD repeat-containing protein 1Aタンパク質・ペプチド : Cullin-1タンパク質・ペプチド : Ubiquitin-conjugating enzyme E2 D2タンパク質・ペプチド : Polyubiquitin-C複合体 : Synthetic peptideタンパク質・ペプチド : CYS-LYS-LYS-ALA-ARG-HIS-ASP-SEP-GLYリガンド : ZINC ION / / / / / / / / / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Pan troglodytes (チンパンジー) / Homo sapiens (ヒト)手法 / / 解像度 : 3.7 Å Baek K / Prabu JR / Schulman BA 資金援助 Organization Grant number 国 Max Planck Society
ジャーナル : Nature / 年 : 2020タイトル : NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly.著者 : Kheewoong Baek / David T Krist / J Rajan Prabu / Spencer Hill / Maren Klügel / Lisa-Marie Neumaier / Susanne von Gronau / Gary Kleiger / Brenda A Schulman / 要旨 : Eukaryotic cell biology depends on cullin-RING E3 ligase (CRL)-catalysed protein ubiquitylation, which is tightly controlled by the modification of cullin with the ubiquitin-like protein NEDD8. ... Eukaryotic cell biology depends on cullin-RING E3 ligase (CRL)-catalysed protein ubiquitylation, which is tightly controlled by the modification of cullin with the ubiquitin-like protein NEDD8. However, how CRLs catalyse ubiquitylation, and the basis of NEDD8 activation, remain unknown. Here we report the cryo-electron microscopy structure of a chemically trapped complex that represents the ubiquitylation intermediate, in which the neddylated CRL1 promotes the transfer of ubiquitin from the E2 ubiquitin-conjugating enzyme UBE2D to its recruited substrate, phosphorylated IκBα. NEDD8 acts as a nexus that binds disparate cullin elements and the RING-activated ubiquitin-linked UBE2D. Local structural remodelling of NEDD8 and large-scale movements of CRL domains converge to juxtapose the substrate and the ubiquitylation active site. These findings explain how a distinctive ubiquitin-like protein alters the functions of its targets, and show how numerous NEDD8-dependent interprotein interactions and conformational changes synergistically configure a catalytic CRL architecture that is both robust, to enable rapid ubiquitylation of the substrate, and fragile, to enable the subsequent functions of cullin-RING proteins. 履歴 登録 2019年12月30日 - ヘッダ(付随情報) 公開 2020年2月12日 - マップ公開 2020年2月12日 - 更新 2024年10月23日 - 現状 2024年10月23日 処理サイト : PDBe / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報
 Homo sapiens (ヒト)
Homo sapiens (ヒト) データ登録者
データ登録者 ドイツ, 1件
ドイツ, 1件  引用
引用 ジャーナル: Nature / 年: 2020
ジャーナル: Nature / 年: 2020

 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_10585.map.gz
emd_10585.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-10585-v30.xml
emd-10585-v30.xml emd-10585.xml
emd-10585.xml EMDBヘッダ
EMDBヘッダ emd_10585_fsc.xml
emd_10585_fsc.xml FSCデータファイル
FSCデータファイル emd_10585.png
emd_10585.png emd_10585_msk_1.map
emd_10585_msk_1.map マスクマップ
マスクマップ emd-10585.cif.gz
emd-10585.cif.gz emd_10585_additional.map.gz
emd_10585_additional.map.gz emd_10585_half_map_1.map.gz
emd_10585_half_map_1.map.gz emd_10585_half_map_2.map.gz
emd_10585_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-10585
http://ftp.pdbj.org/pub/emdb/structures/EMD-10585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10585
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10585 emd_10585_validation.pdf.gz
emd_10585_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_10585_full_validation.pdf.gz
emd_10585_full_validation.pdf.gz emd_10585_validation.xml.gz
emd_10585_validation.xml.gz emd_10585_validation.cif.gz
emd_10585_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10585
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10585 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10585
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10585 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_10585.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_10585.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) emd_10585_msk_1.map
emd_10585_msk_1.map 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 ムービー
ムービー コントローラー
コントローラー








































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)














































 Trichoplusia ni (イラクサキンウワバ)
Trichoplusia ni (イラクサキンウワバ)