+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0202 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the ribosome-NatA complex | |||||||||
 Map data Map data | Ribosome-NatA complex aligned on the ribosome and filtered according to local resolution. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | N-terminal acetylation / protein modification / ribosome / expansion segments / TRANSLATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationN-terminal methionine Nalpha-acetyltransferase NatE / protein-N-terminal-glutamate acetyltransferase activity / N-terminal amino-acid Nalpha-acetyltransferase NatA / NatA complex / protein N-terminal-methionine acetyltransferase activity / protein N-terminal-serine acetyltransferase activity / protein-N-terminal-alanine acetyltransferase activity / protein-N-terminal amino-acid acetyltransferase activity / acetyltransferase activator activity / pre-mRNA 5'-splice site binding ...N-terminal methionine Nalpha-acetyltransferase NatE / protein-N-terminal-glutamate acetyltransferase activity / N-terminal amino-acid Nalpha-acetyltransferase NatA / NatA complex / protein N-terminal-methionine acetyltransferase activity / protein N-terminal-serine acetyltransferase activity / protein-N-terminal-alanine acetyltransferase activity / protein-N-terminal amino-acid acetyltransferase activity / acetyltransferase activator activity / pre-mRNA 5'-splice site binding / cytosolic large ribosomal subunit assembly / mitotic sister chromatid cohesion / response to cycloheximide / cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / SRP-dependent cotranslational protein targeting to membrane / GTP hydrolysis and joining of the 60S ribosomal subunit / negative regulation of mRNA splicing, via spliceosome / preribosome, large subunit precursor / Formation of a pool of free 40S subunits / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / L13a-mediated translational silencing of Ceruloplasmin expression / ribosomal large subunit export from nucleus / translational elongation / translational termination / regulation of translational fidelity / protein-RNA complex assembly / maturation of LSU-rRNA / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / macroautophagy / translational initiation / maintenance of translational fidelity / modification-dependent protein catabolic process / protein tag activity / rRNA processing / ribosome biogenesis / ribosome binding / ribosomal large subunit assembly / 5S rRNA binding / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / negative regulation of translation / rRNA binding / structural constituent of ribosome / protein ubiquitination / ribosome / translation / response to antibiotic / mRNA binding / ubiquitin protein ligase binding / nucleolus / mitochondrion / RNA binding / zinc ion binding / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Knorr AG / Becker T / Berninghausen O / Beckmann R | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation. Authors: Alexandra G Knorr / Christian Schmidt / Petr Tesina / Otto Berninghausen / Thomas Becker / Birgitta Beatrix / Roland Beckmann /  Abstract: The majority of eukaryotic proteins are N-terminally α-acetylated by N-terminal acetyltransferases (NATs). Acetylation usually occurs co-translationally and defects have severe consequences. ...The majority of eukaryotic proteins are N-terminally α-acetylated by N-terminal acetyltransferases (NATs). Acetylation usually occurs co-translationally and defects have severe consequences. Nevertheless, it is unclear how these enzymes act in concert with the translating ribosome. Here, we report the structure of a native ribosome-NatA complex from Saccharomyces cerevisiae. NatA (comprising Naa10, Naa15 and Naa50) displays a unique mode of ribosome interaction by contacting eukaryotic-specific ribosomal RNA expansion segments in three out of four binding patches. Thereby, NatA is dynamically positioned directly underneath the ribosomal exit tunnel to facilitate modification of the emerging nascent peptide chain. Methionine amino peptidases, but not chaperones or signal recognition particle, would be able to bind concomitantly. This work assigns a function to the hitherto enigmatic ribosomal RNA expansion segments and provides mechanistic insights into co-translational protein maturation by N-terminal acetylation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0202.map.gz emd_0202.map.gz | 162.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0202-v30.xml emd-0202-v30.xml emd-0202.xml emd-0202.xml | 78.4 KB 78.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0202.png emd_0202.png | 192.9 KB | ||
| Filedesc metadata |  emd-0202.cif.gz emd-0202.cif.gz | 15.2 KB | ||
| Others |  emd_0202_additional.map.gz emd_0202_additional.map.gz | 264.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0202 http://ftp.pdbj.org/pub/emdb/structures/EMD-0202 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0202 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0202 | HTTPS FTP |
-Related structure data
| Related structure data |  6hd7MC  0201C  0203C  6hd5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0202.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0202.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
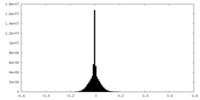
| Annotation | Ribosome-NatA complex aligned on the ribosome and filtered according to local resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.084 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Ribosome-NatA complex aligned on the ribosome, postprocessed unfiltered...
| File | emd_0202_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
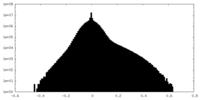
| Annotation | Ribosome-NatA complex aligned on the ribosome, postprocessed unfiltered map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Ribosome-NatA complex
+Supramolecule #1: Ribosome-NatA complex
+Supramolecule #2: 60S ribosomal subunit
+Supramolecule #3: N-terminal acetyl transferase A
+Macromolecule #1: Saccharomyces cerevisiae S288C 35S pre-ribosomal RNA (RDN37-1), m...
+Macromolecule #2: 5S rRNA
+Macromolecule #3: 5.8S rRNA
+Macromolecule #4: tRNA
+Macromolecule #5: P-site tRNA
+Macromolecule #6: 60S ribosomal protein L42-A
+Macromolecule #7: 60S ribosomal protein L43-A
+Macromolecule #8: 60S ribosomal protein L2-A
+Macromolecule #9: 60S ribosomal protein L3
+Macromolecule #10: 60S ribosomal protein L4-A
+Macromolecule #11: 60S ribosomal protein L5
+Macromolecule #12: 60S ribosomal protein L6-A
+Macromolecule #13: 60S ribosomal protein L7-A
+Macromolecule #14: 60S ribosomal protein L8-A
+Macromolecule #15: 60S ribosomal protein L9-A
+Macromolecule #16: 60S ribosomal protein L11-A
+Macromolecule #17: 60S ribosomal protein L13-A
+Macromolecule #18: 60S ribosomal protein L14-A
+Macromolecule #19: 60S ribosomal protein L15-A
+Macromolecule #20: 60S ribosomal protein L16-A
+Macromolecule #21: 60S ribosomal protein L17-A
+Macromolecule #22: 60S ribosomal protein L18-A
+Macromolecule #23: 60S ribosomal protein L19-A
+Macromolecule #24: 60S ribosomal protein L20-A
+Macromolecule #25: 60S ribosomal protein L21-A
+Macromolecule #26: 60S ribosomal protein L22-A
+Macromolecule #27: 60S ribosomal protein L23-A
+Macromolecule #28: 60S ribosomal protein L24-A
+Macromolecule #29: 60S ribosomal protein L25
+Macromolecule #30: 60S ribosomal protein L26-A
+Macromolecule #31: 60S ribosomal protein L27-A
+Macromolecule #32: 60S ribosomal protein L28
+Macromolecule #33: 60S ribosomal protein L29
+Macromolecule #34: 60S ribosomal protein L30
+Macromolecule #35: 60S ribosomal protein L31-A
+Macromolecule #36: 60S ribosomal protein L32
+Macromolecule #37: 60S ribosomal protein L33-A
+Macromolecule #38: 60S ribosomal protein L34-A
+Macromolecule #39: 60S ribosomal protein L35-A
+Macromolecule #40: 60S ribosomal protein L36-A
+Macromolecule #41: 60S ribosomal protein L37-A
+Macromolecule #42: 60S ribosomal protein L38
+Macromolecule #43: 60S ribosomal protein L39
+Macromolecule #44: Ubiquitin-60S ribosomal protein L40
+Macromolecule #45: 60S ribosomal protein L41-A
+Macromolecule #46: ribosomal protein RPL1
+Macromolecule #47: 60S ribosomal protein L10
+Macromolecule #48: N-terminal acetyltransferase A complex subunit NAT1
+Macromolecule #49: N-terminal acetyltransferase A complex catalytic subunit ARD1
+Macromolecule #50: N-alpha-acetyltransferase NAT5
+Macromolecule #51: nascent polypeptide chain
+Macromolecule #52: 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethy...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 2.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6hd7: |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)