+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0188 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Echovirus 18 Open particle without three pentamers | |||||||||
 Map data Map data | Echovirus 18 Open particle without three pentamers | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | echovirus / echovirus 18 / open particle / O-particle / enterovirus / picornavirus / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Echovirus E18 Echovirus E18 | |||||||||
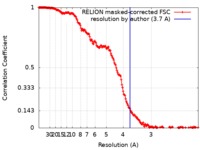
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Buchta D / Fuzik T | |||||||||
| Funding support |  Czech Republic, 1 items Czech Republic, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Enterovirus particles expel capsid pentamers to enable genome release. Authors: David Buchta / Tibor Füzik / Dominik Hrebík / Yevgen Levdansky / Lukáš Sukeník / Liya Mukhamedova / Jana Moravcová / Robert Vácha / Pavel Plevka /   Abstract: Viruses from the genus Enterovirus are important human pathogens. Receptor binding or exposure to acidic pH in endosomes converts enterovirus particles to an activated state that is required for ...Viruses from the genus Enterovirus are important human pathogens. Receptor binding or exposure to acidic pH in endosomes converts enterovirus particles to an activated state that is required for genome release. However, the mechanism of enterovirus uncoating is not well understood. Here, we use cryo-electron microscopy to visualize virions of human echovirus 18 in the process of genome release. We discover that the exit of the RNA from the particle of echovirus 18 results in a loss of one, two, or three adjacent capsid-protein pentamers. The opening in the capsid, which is more than 120 Å in diameter, enables the release of the genome without the need to unwind its putative double-stranded RNA segments. We also detect capsids lacking pentamers during genome release from echovirus 30. Thus, our findings uncover a mechanism of enterovirus genome release that could become target for antiviral drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0188.map.gz emd_0188.map.gz | 41.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0188-v30.xml emd-0188-v30.xml emd-0188.xml emd-0188.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0188_fsc.xml emd_0188_fsc.xml | 18.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0188.png emd_0188.png | 201.7 KB | ||
| Masks |  emd_0188_msk_1.map emd_0188_msk_1.map emd_0188_msk_2.map emd_0188_msk_2.map | 512 MB 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0188.cif.gz emd-0188.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0188 http://ftp.pdbj.org/pub/emdb/structures/EMD-0188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0188 | HTTPS FTP |
-Validation report
| Summary document |  emd_0188_validation.pdf.gz emd_0188_validation.pdf.gz | 552.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0188_full_validation.pdf.gz emd_0188_full_validation.pdf.gz | 551.9 KB | Display | |
| Data in XML |  emd_0188_validation.xml.gz emd_0188_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_0188_validation.cif.gz emd_0188_validation.cif.gz | 22.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0188 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0188 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0188 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0188 | HTTPS FTP |
-Related structure data
| Related structure data |  6hblMC  0181C  0182C  0183C  0184C  0185C  0186C  0187C  0189C  0217C  6hbgC  6hbhC  6hbjC  6hbkC  6hhtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0188.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0188.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Echovirus 18 Open particle without three pentamers | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.061 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0188_msk_1.map emd_0188_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_0188_msk_2.map emd_0188_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Echovirus E18
| Entire | Name:  Echovirus E18 Echovirus E18 |
|---|---|
| Components |
|
-Supramolecule #1: Echovirus E18
| Supramolecule | Name: Echovirus E18 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 47506 / Sci species name: Echovirus E18 / Sci species strain: Metcalf / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.33 MDa |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 340.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Echovirus 18 capsid protein 1
| Macromolecule | Name: Echovirus 18 capsid protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Echovirus E18 Echovirus E18 |
| Molecular weight | Theoretical: 32.564445 KDa |
| Sequence | String: GDNQDRTVAN TQPSGPSNST EIPALTAVET GHTSQVDPSD TIQTRHVVNF HSRSESTIEN FMGRAACVFM DQYKINGEET STDRFAVWT INIREMAQLR RKCEMFTYMR FDIEMTMVIT SCQDQGTILD QDMPVLTHQI MYVPPGGPIP AKVDGYEWQT S TNPSVFWT ...String: GDNQDRTVAN TQPSGPSNST EIPALTAVET GHTSQVDPSD TIQTRHVVNF HSRSESTIEN FMGRAACVFM DQYKINGEET STDRFAVWT INIREMAQLR RKCEMFTYMR FDIEMTMVIT SCQDQGTILD QDMPVLTHQI MYVPPGGPIP AKVDGYEWQT S TNPSVFWT EGNAPPRISI PFISVGNAYS SFYDGWSHFT QDGTYGYTTL NAMGKLYIRH VNRSSPHQIT STIRVYFKPK HI KAWVPRP PRLCPYINKR DVNFVVTEIT DSRTSITDTP HPEHSVLATH UniProtKB: Genome polyprotein |
-Macromolecule #2: Echovirus 18 capsid protein 2
| Macromolecule | Name: Echovirus 18 capsid protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Echovirus E18 Echovirus E18 |
| Molecular weight | Theoretical: 28.802328 KDa |
| Sequence | String: SPSAEECGYS DRVRSMTLGN STITTQESAN VVVGYGEWPS YLSDREATAE DQPTQPDVAT CRFYTLESVQ WEKTSPGWWW KFPEALKNM GLFGQNMHYH YLGRAGYTIH VQCNASKFHQ GCLLVVCVPE AEMGCADTDT TFPATELTTE DTPHVFTSDS I TGKKVQAA ...String: SPSAEECGYS DRVRSMTLGN STITTQESAN VVVGYGEWPS YLSDREATAE DQPTQPDVAT CRFYTLESVQ WEKTSPGWWW KFPEALKNM GLFGQNMHYH YLGRAGYTIH VQCNASKFHQ GCLLVVCVPE AEMGCADTDT TFPATELTTE DTPHVFTSDS I TGKKVQAA VCNAGMGVGV GNLTIFPHQW INLRTNNSAT IVIPYINSVP MDNMFRHYNF TLMIIPFAPL NFTDGATAYV PI TVTIAPM YAEYNGLRLA STQ UniProtKB: Genome polyprotein |
-Macromolecule #3: Echovirus 18 capsid protein 3
| Macromolecule | Name: Echovirus 18 capsid protein 3 / type: protein_or_peptide / ID: 3 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Echovirus E18 Echovirus E18 |
| Molecular weight | Theoretical: 26.143783 KDa |
| Sequence | String: GVPVLNTPGS NQFLTSDDYQ SPSAMPQFDE TPEMHIPGEV RNLMEIAEVD SVVPVNNVTG KTKSMDAYQI PVGTGNTDKT KPIFSFQMD PGYSSVLKRT LLGEMLNYYA HWSGSVKLTF LFCGSAMATG KLLISYSPPG ASVPTSRKDA MLGTHIVWDI G LQSSCVLC ...String: GVPVLNTPGS NQFLTSDDYQ SPSAMPQFDE TPEMHIPGEV RNLMEIAEVD SVVPVNNVTG KTKSMDAYQI PVGTGNTDKT KPIFSFQMD PGYSSVLKRT LLGEMLNYYA HWSGSVKLTF LFCGSAMATG KLLISYSPPG ASVPTSRKDA MLGTHIVWDI G LQSSCVLC VPWISQSHYR MVQQDPYTSA GYITCWYQTN IVVPPGAPTS CDVLCFASAC NDFSVRLLRD TPFMAQPGKL Q UniProtKB: Genome polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: FORMVAR / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 6331 / Average exposure time: 1.0 sec. / Average electron dose: 45.2 e/Å2 Details: Images were collected in movie-mode at 39 frames per second |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 3.282 µm / Calibrated defocus min: 0.651 µm / Calibrated magnification: 79575 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)