+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0184 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Echovirus 18 Open particle without one pentamer | |||||||||
 Map data Map data | Echovirus 18 Open particle without one pentamer with C1 symmetry | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Echovirus E18 Echovirus E18 | |||||||||
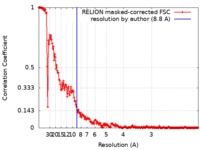
| Method | single particle reconstruction / cryo EM / Resolution: 8.8 Å | |||||||||
 Authors Authors | Buchta D / Fuzik T / Hrebik D / Levdansky Y / Moravcova J / Plevka P | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Enterovirus particles expel capsid pentamers to enable genome release. Authors: David Buchta / Tibor Füzik / Dominik Hrebík / Yevgen Levdansky / Lukáš Sukeník / Liya Mukhamedova / Jana Moravcová / Robert Vácha / Pavel Plevka /   Abstract: Viruses from the genus Enterovirus are important human pathogens. Receptor binding or exposure to acidic pH in endosomes converts enterovirus particles to an activated state that is required for ...Viruses from the genus Enterovirus are important human pathogens. Receptor binding or exposure to acidic pH in endosomes converts enterovirus particles to an activated state that is required for genome release. However, the mechanism of enterovirus uncoating is not well understood. Here, we use cryo-electron microscopy to visualize virions of human echovirus 18 in the process of genome release. We discover that the exit of the RNA from the particle of echovirus 18 results in a loss of one, two, or three adjacent capsid-protein pentamers. The opening in the capsid, which is more than 120 Å in diameter, enables the release of the genome without the need to unwind its putative double-stranded RNA segments. We also detect capsids lacking pentamers during genome release from echovirus 30. Thus, our findings uncover a mechanism of enterovirus genome release that could become target for antiviral drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0184.map.gz emd_0184.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0184-v30.xml emd-0184-v30.xml emd-0184.xml emd-0184.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0184_fsc.xml emd_0184_fsc.xml | 17.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_0184.png emd_0184.png | 237.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0184 http://ftp.pdbj.org/pub/emdb/structures/EMD-0184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0184 | HTTPS FTP |
-Related structure data
| Related structure data |  0181C  0182C  0183C  0185C  0186C  0187C  0188C  0189C  0217C  6hbgC  6hbhC  6hbjC  6hbkC  6hblC  6hhtC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0184.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0184.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Echovirus 18 Open particle without one pentamer with C1 symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Echovirus E18
| Entire | Name:  Echovirus E18 Echovirus E18 |
|---|---|
| Components |
|
-Supramolecule #1: Echovirus E18
| Supramolecule | Name: Echovirus E18 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 47506 / Sci species name: Echovirus E18 / Sci species strain: Metcalf / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.20 MDa |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 340.0 Å / T number (triangulation number): 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: FORMVAR / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 14.0 µm / Number grids imaged: 1 / Number real images: 6331 / Average exposure time: 1.0 sec. / Average electron dose: 45.2 e/Å2 Details: Images were collected in movie-mode at 39 frames per second |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 3.282 µm / Calibrated defocus min: 0.651 µm / Calibrated magnification: 79725 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Overall B value: 223.5 |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)