+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-tomography and subtomogram averaging of Sar1-Sec23-Sec24. | |||||||||
 Map data Map data | Map sharpened with B-factor -350 and locally filtered with LocRes in RELION. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | COPII coat / membrane trafficking / protein transport | |||||||||
| Function / homology |  Function and homology information Function and homology informationAntigen Presentation: Folding, assembly and peptide loading of class I MHC / Cargo concentration in the ER / regulation of COPII vesicle coating / positive regulation of ER to Golgi vesicle-mediated transport / nuclear envelope organization / COPII-mediated vesicle transport / mitochondria-associated endoplasmic reticulum membrane contact site / COPII-coated vesicle cargo loading / positive regulation of protein exit from endoplasmic reticulum / vesicle organization ...Antigen Presentation: Folding, assembly and peptide loading of class I MHC / Cargo concentration in the ER / regulation of COPII vesicle coating / positive regulation of ER to Golgi vesicle-mediated transport / nuclear envelope organization / COPII-mediated vesicle transport / mitochondria-associated endoplasmic reticulum membrane contact site / COPII-coated vesicle cargo loading / positive regulation of protein exit from endoplasmic reticulum / vesicle organization / COPII vesicle coat / signal sequence binding / membrane organization / mitochondrial fission / fungal-type vacuole membrane / reticulophagy / mitochondrial membrane organization / endoplasmic reticulum exit site / endoplasmic reticulum to Golgi vesicle-mediated transport / GTPase activator activity / SNARE binding / macroautophagy / intracellular protein transport / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Golgi membrane / GTPase activity / endoplasmic reticulum membrane / GTP binding / endoplasmic reticulum / mitochondrion / zinc ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Hutchings J / Zanetti G | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Subtomogram averaging of COPII assemblies reveals how coat organization dictates membrane shape. Authors: Joshua Hutchings / Viktoriya Stancheva / Elizabeth A Miller / Giulia Zanetti /  Abstract: Eukaryotic cells employ membrane-bound carriers to transport cargo between compartments in a process essential to cell functionality. Carriers are generated by coat complexes that couple cargo ...Eukaryotic cells employ membrane-bound carriers to transport cargo between compartments in a process essential to cell functionality. Carriers are generated by coat complexes that couple cargo capture to membrane deformation. The COPII coat mediates export from the endoplasmic reticulum by assembling in inner and outer layers, yielding carriers of variable shape and size that allow secretion of thousands of diverse cargo. Despite detailed understanding of COPII subunits, the molecular mechanisms of coat assembly and membrane deformation are unclear. Here we present a 4.9 Å cryo-tomography subtomogram averaging structure of in vitro-reconstituted membrane-bound inner coat. We show that the outer coat (Sec13-Sec31) bridges inner coat subunits (Sar1-Sec23-Sec24), promoting their assembly into a tight lattice. We directly visualize the membrane-embedded Sar1 amphipathic helix, revealing that lattice formation induces parallel helix insertions, yielding tubular curvature. We propose that regulators like the procollagen receptor TANGO1 modulate this mechanism to determine vesicle shape and size. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0044.map.gz emd_0044.map.gz | 25.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0044-v30.xml emd-0044-v30.xml emd-0044.xml emd-0044.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
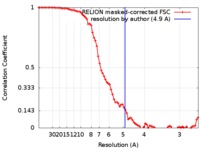
| FSC (resolution estimation) |  emd_0044_fsc.xml emd_0044_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0044.png emd_0044.png | 518.3 KB | ||
| Masks |  emd_0044_msk_1.map emd_0044_msk_1.map emd_0044_msk_2.map emd_0044_msk_2.map | 42.9 MB 42.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0044.cif.gz emd-0044.cif.gz | 6.7 KB | ||
| Others |  emd_0044_half_map_1.map.gz emd_0044_half_map_1.map.gz emd_0044_half_map_2.map.gz emd_0044_half_map_2.map.gz | 21.8 MB 21.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0044 http://ftp.pdbj.org/pub/emdb/structures/EMD-0044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0044 | HTTPS FTP |
-Related structure data
| Related structure data |  6gniMC  0307C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0044.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0044.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map sharpened with B-factor -350 and locally filtered with LocRes in RELION. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.327 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_0044_msk_1.map emd_0044_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_0044_msk_2.map emd_0044_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Average map from independently aligned half dataset 2.
| File | emd_0044_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Average map from independently aligned half dataset 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Average map from independently aligned half dataset 1.
| File | emd_0044_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Average map from independently aligned half dataset 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : COPII coat assembled on lipid bilayer
| Entire | Name: COPII coat assembled on lipid bilayer |
|---|---|
| Components |
|
-Supramolecule #1: COPII coat assembled on lipid bilayer
| Supramolecule | Name: COPII coat assembled on lipid bilayer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Protein transport protein SEC23
| Macromolecule | Name: Protein transport protein SEC23 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 85.332047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DFETNEDING VRFTWNVFPS TRSDANSNVV PVGCLYTPLK EYDELNVAPY NPVVCSGPHC KSILNPYCVI DPRNSSWSCP ICNSRNHLP PQYTNLSQEN MPLELQSTTI EYITNKPVTV PPIFFFVVDL TSETENLDSL KESIITSLSL LPPNALIGLI T YGNVVQLH ...String: DFETNEDING VRFTWNVFPS TRSDANSNVV PVGCLYTPLK EYDELNVAPY NPVVCSGPHC KSILNPYCVI DPRNSSWSCP ICNSRNHLP PQYTNLSQEN MPLELQSTTI EYITNKPVTV PPIFFFVVDL TSETENLDSL KESIITSLSL LPPNALIGLI T YGNVVQLH DLSSETIDRC NVFRGDREYQ LEALTEMLTG QKPTGPGGAA SHLPNAMNKV TPFSLNRFFL PLEQVEFKLN QL LENLSPD QWSVPAGHRP LRATGSALNI ASLLLQGCYK NIPARIILFA SGPGTVAPGL IVNSELKDPL RSHHDIDSDH AQH YKKACK FYNQIAQRVA ANGHTVDIFA GCYDQIGMSE MKQLTDSTGG VLLLTDAFST AIFKQSYLRL FAKDEEGYLK MAFN GNMAV KTSKDLKVQG LIGHASAVKK TDANNISESE IGIGATSTWK MASLSPYHSY AIFFEIANTA ANSNPMMSAP GSADR PHLA YTQFITTYQH SSGTNRIRVT TVANQLLPFG TPAIAASFDQ EAAAVLMARI AVHKAETDDG ADVIRWLDRT LIKLCQ KYA DYNKDDPQSF RLAPNFSLYP QFTYYLRRSQ FLSVFNNSPD ETAFYRHIFT REDTTNSLIM IQPTLTSFSM EDDPQPV LL DSISVKPNTI LLLDTFFFIL IYHGEQIAQW RKAGYQDDPQ YADFKALLEE PKLEAAELLV DRFPLPRFID TEAGGSQA R FLLSKLNPSD NYQDMARGGS TIVLTDDVSL QNFMTHLQQV AVSGQA UniProtKB: Protein transport protein SEC23 |
-Macromolecule #2: Protein transport protein SEC24
| Macromolecule | Name: Protein transport protein SEC24 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 89.294953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: RPMNQLYPID LLTELPPPIT DLTLPPPPLV IPPERMLVPS ELSNASPDYI RSTLNAVPKN SSLLKKSKLP FGLVIRPYQH LYDDIDPPP LNEDGLIVRC RRCRSYMNPF VTFIEQGRRW RCNFCRLAND VPMQMDQSDP NDPKSRYDRN EIKCAVMEYM A PKEYTLRQ ...String: RPMNQLYPID LLTELPPPIT DLTLPPPPLV IPPERMLVPS ELSNASPDYI RSTLNAVPKN SSLLKKSKLP FGLVIRPYQH LYDDIDPPP LNEDGLIVRC RRCRSYMNPF VTFIEQGRRW RCNFCRLAND VPMQMDQSDP NDPKSRYDRN EIKCAVMEYM A PKEYTLRQ PPPATYCFLI DVSQSSIKSG LLATTINTLL QNLDSIPNHD ERTRISILCV DNAIHYFKIP LDSENNEESA DQ INMMDIA DLEEPFLPRP NSMVVSLKAC RQNIETLLTK IPQIFQSNLI TNFALGPALK SAYHLIGGVG GKIIVVSGTL PNL GIGKLQ RRNESGVVNT SKETAQLLSC QDSFYKNFTI DCSKVQITVD LFLASEDYMD VASLSNLSRF TAGQTHFYPG FSGK NPNDI VKFSTEFAKH ISMDFCMETV MRARGSTGLR MSRFYGHFFN RSSDLCAFST MPRDQSYLFE VNVDESIMAD YCYVQ VAVL LSLNNSQRRI RIITLAMPTT ESLAEVYASA DQLAIASFYN SKAVEKALNS SLDDARVLIN KSVQDILATY KKEIVV SNT AGGAPLRLCA NLRMFPLLMH SLTKHMAFRS GIVPSDHRAS ALNNLESLPL KYLIKNIYPD VYSLHDMADE AGLPVQT ED GEATGTIVLP QPINATSSLF ERYGLYLIDN GNELFLWMGG DAVPALVFDV FGTQDIFDIP IGKQEIPVVE NSEFNQRV R NIINQLRNHD DVITYQSLYI VRGASLSEPV NHASAREVAT LRLWASSTLV EDKILNNESY REFLQIMKAR ISK UniProtKB: Protein transport protein SEC24 |
-Macromolecule #3: Small COPII coat GTPase SAR1
| Macromolecule | Name: Small COPII coat GTPase SAR1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.79242 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HGKLLFLGLD NAGKTTLLHM LKNDRLATLQ PTWHPTSEEL AIGNIKFTTF DLGGHIQARR LWKDYFPEVN GIVFLVDAAD PERFDEARV ELDALFNIAE LKDVPFVILG NKIDAPNAVS EAELRSALGL LNTTGSQRIE GQRPVEVFMC SVVMRNGYLE A FQWLSQY UniProtKB: Small COPII coat GTPase SAR1 |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER / type: ligand / ID: 5 / Number of copies: 1 / Formula: GNP |
|---|---|
| Molecular weight | Theoretical: 522.196 Da |
| Chemical component information |  ChemComp-GNP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 3.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6gni: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)