+Search query
-Structure paper
| Title | Delta-type glutamate receptors are ligand-gated ion channels. |
|---|---|
| Journal, issue, pages | Nature, Vol. 647, Issue 8091, Page 1063-1071, Year 2025 |
| Publish date | Sep 16, 2025 |
 Authors Authors | Haobo Wang / Fairine Ahmed / Jeffrey Khau / Anish Kumar Mondal / Edward C Twomey /  |
| PubMed Abstract | Delta-type ionotropic glutamate receptors (iGluRs, also known as GluDs) are members of the iGluR ligand-gated ion channel family, yet their function remains unknown. Although GluDs are widely ...Delta-type ionotropic glutamate receptors (iGluRs, also known as GluDs) are members of the iGluR ligand-gated ion channel family, yet their function remains unknown. Although GluDs are widely expressed in the brain, have key roles in synaptic organization, and harbour disease-linked mutations, whether they retain iGluR-like channel function is debated as currents have not been directly observed. Here we define GluDs as ligand-gated ion channels that are tightly regulated in cellular contexts by purifying human GluD2 (hGluD2) and directly characterizing its structure and function using cryo-electron microscopy and bilayer recordings. We show that hGluD2 is activated by D-serine and GABA (γ-aminobutyric acid), with augmented activation at physiological temperatures. We reveal that hGluD2 contains an ion channel directly coupled to clamshell-like ligand-binding domains, which are coordinated by the amino-terminal domain above the ion channel. Ligand binding triggers channel opening via an asymmetric mechanism, and a cerebellar ataxia point mutation in the ligand-binding domain rearranges the receptor architecture and induces leak currents. Our findings demonstrate that GluDs possess the intrinsic biophysical properties of ligand-gated ion channels, reconciling prior conflicting observations to establish a framework for understanding their cellular regulation and for developing therapies targeting GluD2. |
 External links External links |  Nature / Nature /  PubMed:40957579 / PubMed:40957579 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.57 - 3.74 Å |
| Structure data | EMDB-49888, PDB-9nwo: EMDB-49889, PDB-9nwp: EMDB-49890, PDB-9nwq: EMDB-70667, PDB-9ooo: EMDB-70668, PDB-9oop: |
| Chemicals | 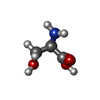 ChemComp-DSN: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN / Ligand-gated ion channel / ion channel / neurotransmitter receptor |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers













 homo sapiens (human)
homo sapiens (human)