+Search query
-Structure paper
| Title | Structural basis for synthase activation and cellulose modification in the E. coli Type II Bcs secretion system. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 15, Issue 1, Page 8799, Year 2024 |
| Publish date | Oct 11, 2024 |
 Authors Authors | Itxaso Anso / Samira Zouhir / Thibault Géry Sana / Petya Violinova Krasteva /   |
| PubMed Abstract | Bacterial cellulosic polymers constitute a prevalent class of biofilm matrix exopolysaccharides that are synthesized by several types of bacterial cellulose secretion (Bcs) systems, which include ...Bacterial cellulosic polymers constitute a prevalent class of biofilm matrix exopolysaccharides that are synthesized by several types of bacterial cellulose secretion (Bcs) systems, which include conserved cyclic diguanylate (c-di-GMP)-dependent cellulose synthase modules together with diverse accessory subunits. In E. coli, the biogenesis of phosphoethanolamine (pEtN)-modified cellulose relies on the BcsRQABEFG macrocomplex, encompassing inner-membrane and cytosolic subunits, and an outer membrane porin, BcsC. Here, we use cryogenic electron microscopy to shed light on the molecular mechanisms of BcsA-dependent recruitment and stabilization of a trimeric BcsG pEtN-transferase for polymer modification, and a dimeric BcsF-dependent recruitment of an otherwise cytosolic BcsERQ regulatory complex. We further demonstrate that BcsE, a secondary c-di-GMP sensor, can remain dinucleotide-bound and retain the essential-for-secretion BcsRQ partners onto the synthase even in the absence of direct c-di-GMP-synthase complexation, likely lowering the threshold for c-di-GMP-dependent synthase activation. Such activation-by-proxy mechanism could allow Bcs secretion system activity even in the absence of substantial intracellular c-di-GMP increase, and is reminiscent of other widespread synthase-dependent polysaccharide secretion systems where dinucleotide sensing and/or synthase stabilization are carried out by key co-polymerase subunits. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:39394223 / PubMed:39394223 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.35 - 7.0 Å |
| Structure data | EMDB-50567, PDB-9fmt: EMDB-50571, PDB-9fmv: EMDB-50581, PDB-9fmz:  EMDB-50584: Cryo-EM structure of the c-di-GMP-saturated Bcs macrocomplex (BcsABR2Q2E2F2G3) for cellulose secretion of E. coli (filtered to 6A)  EMDB-50595: Cryo-EM structure of the c-di-GMP non-saturated Bcs macrocomplex (BcsABR2Q2E2F2G3) for cellulose secretion of E. coli (filtered to 7A) EMDB-50599, PDB-9fnn: EMDB-50619, PDB-9fo7: EMDB-50632, PDB-9fp0: EMDB-50633, PDB-9fp2: |
| Chemicals | 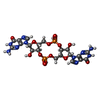 ChemComp-C2E:  ChemComp-ATP:  ChemComp-MG: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Bacterial cellulose secretion |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers


















