[English] 日本語
 Yorodumi
Yorodumi- EMDB-50633: Cryo-EM structure of the BcsEFRQ regulatory subcomplex for E. col... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the BcsEFRQ regulatory subcomplex for E. coli cellulose secretion in non-saturating c-di-GMP (local) | |||||||||
 Map data Map data | Deep Emhancer-sharpened map of the BcsE2F2 regulatory subcomplex from the E. coli cellulose secretion system in non-saturating c-di-GMP (local) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacterial cellulose secretion / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of cell division / cytoplasmic side of plasma membrane / cell division / ATP hydrolysis activity / ATP binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.76 Å | |||||||||
 Authors Authors | Anso I / Krasteva PV | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for synthase activation and cellulose modification in the E. coli Type II Bcs secretion system. Authors: Itxaso Anso / Samira Zouhir / Thibault Géry Sana / Petya Violinova Krasteva /   Abstract: Bacterial cellulosic polymers constitute a prevalent class of biofilm matrix exopolysaccharides that are synthesized by several types of bacterial cellulose secretion (Bcs) systems, which include ...Bacterial cellulosic polymers constitute a prevalent class of biofilm matrix exopolysaccharides that are synthesized by several types of bacterial cellulose secretion (Bcs) systems, which include conserved cyclic diguanylate (c-di-GMP)-dependent cellulose synthase modules together with diverse accessory subunits. In E. coli, the biogenesis of phosphoethanolamine (pEtN)-modified cellulose relies on the BcsRQABEFG macrocomplex, encompassing inner-membrane and cytosolic subunits, and an outer membrane porin, BcsC. Here, we use cryogenic electron microscopy to shed light on the molecular mechanisms of BcsA-dependent recruitment and stabilization of a trimeric BcsG pEtN-transferase for polymer modification, and a dimeric BcsF-dependent recruitment of an otherwise cytosolic BcsERQ regulatory complex. We further demonstrate that BcsE, a secondary c-di-GMP sensor, can remain dinucleotide-bound and retain the essential-for-secretion BcsRQ partners onto the synthase even in the absence of direct c-di-GMP-synthase complexation, likely lowering the threshold for c-di-GMP-dependent synthase activation. Such activation-by-proxy mechanism could allow Bcs secretion system activity even in the absence of substantial intracellular c-di-GMP increase, and is reminiscent of other widespread synthase-dependent polysaccharide secretion systems where dinucleotide sensing and/or synthase stabilization are carried out by key co-polymerase subunits. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50633.map.gz emd_50633.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50633-v30.xml emd-50633-v30.xml emd-50633.xml emd-50633.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50633_fsc.xml emd_50633_fsc.xml | 16.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_50633.png emd_50633.png | 50.2 KB | ||
| Masks |  emd_50633_msk_1.map emd_50633_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50633.cif.gz emd-50633.cif.gz | 7.6 KB | ||
| Others |  emd_50633_additional_1.map.gz emd_50633_additional_1.map.gz emd_50633_half_map_1.map.gz emd_50633_half_map_1.map.gz emd_50633_half_map_2.map.gz emd_50633_half_map_2.map.gz | 224.8 MB 441.8 MB 441.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50633 http://ftp.pdbj.org/pub/emdb/structures/EMD-50633 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50633 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50633 | HTTPS FTP |
-Related structure data
| Related structure data |  9fp2MC  9fmtC  9fmvC  9fmzC  9fnnC  9fo7C  9fp0C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50633.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50633.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Deep Emhancer-sharpened map of the BcsE2F2 regulatory subcomplex from the E. coli cellulose secretion system in non-saturating c-di-GMP (local) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.839 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50633_msk_1.map emd_50633_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map of the BcsE2F2 regulatory subcomplex from...
| File | emd_50633_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map of the BcsE2F2 regulatory subcomplex from the E. coli cellulose secretion system in non-saturating c-di-GMP (local) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map of the BcsE2F2 regulatory subcomplex from the...
| File | emd_50633_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of the BcsE2F2 regulatory subcomplex from the E. coli cellulose secretion system in non-saturating c-di-GMP (local) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map of the BcsE2F2 regulatory subcomplex from the...
| File | emd_50633_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of the BcsE2F2 regulatory subcomplex from the E. coli cellulose secretion system in non-saturating c-di-GMP (local) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Locally refined BcsEFRQ regulatory subcomplex in non-saturating c...
| Entire | Name: Locally refined BcsEFRQ regulatory subcomplex in non-saturating c-di-GMP from the E. coli cellulose secretion system |
|---|---|
| Components |
|
-Supramolecule #1: Locally refined BcsEFRQ regulatory subcomplex in non-saturating c...
| Supramolecule | Name: Locally refined BcsEFRQ regulatory subcomplex in non-saturating c-di-GMP from the E. coli cellulose secretion system type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Local refinement in the context of an assembled macrocomplex |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 990 KDa |
-Macromolecule #1: Cyclic di-GMP binding protein BcsE
| Macromolecule | Name: Cyclic di-GMP binding protein BcsE / type: protein_or_peptide / ID: 1 Details: Purified Bcs macrocomplex with stoichiometry BcsA-BcsB6-BcsR2-BcsQ2-BcsE2-BcsF2-BcsG3 Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60.967539 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASWSHPQFE KGSMRDIVDP VFSIGISSLW DELRHMPAGG VWWFNVDRHE DAISLANQTI ASQAETAHVA VISMDSDPAK IFQLDDSQG PEKIKLFSML NHEKGLYYLT RDLQCSIDPH NYLFILVCAN NAWQNIPAER LRSWLDKMNK WSRLNHCSLL V INPGNNND ...String: MASWSHPQFE KGSMRDIVDP VFSIGISSLW DELRHMPAGG VWWFNVDRHE DAISLANQTI ASQAETAHVA VISMDSDPAK IFQLDDSQG PEKIKLFSML NHEKGLYYLT RDLQCSIDPH NYLFILVCAN NAWQNIPAER LRSWLDKMNK WSRLNHCSLL V INPGNNND KQFSLLLEEY RSLFGLASLR FQGDQHLLDI AFWCNEKGVS ARQQLSVQQQ NGIWTLVQSE EAEIQPRSDE KR ILSNVAV LEGAPPLSEH WQLFNNNEVL FNEARTAQAA TVVFSLQQNA QIEPLARSIH TLRRQRGSAM KILVRENTAS LRA TDERLL LACGANMVIP WNAPLSRCLT MIESVQGQKF SRYVPEDITT LLSMTQPLKL RGFQKWDVFC NAVNNMMNNP LLPA HGKGV LVALRPVPGI RVEQALTLCR PNRTGDIMTI GGNRLVLFLS FCRINDLDTA LNHIFPLPTG DIFSNRMVWF EDDQI SAEL VQMRLLAPEQ WGMPLPLTQS SKPVINAEHD GRHWRRIPEP MRLLDDAVER SS |
-Macromolecule #2: Cell division protein
| Macromolecule | Name: Cell division protein / type: protein_or_peptide / ID: 2 Details: Purified Bcs macrocomplex with stoichiometry BcsA-BcsB6-BcsR2-BcsQ2-BcsE2-BcsF2-BcsG3 Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.960832 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAVLGLQGVR GGVGTTTITA ALAWSLQMLG ENVLVVDACP DNLLRLSFNV DFTHRQGWAR AMLDGQDWRD AGLRYTSQLD LLPFGQLSI EEQENPQHWQ TRLSDICSGL QQLKASGRYQ WILIDLPRDA SQITHQLLSL CDHSLAIVNV DANCHIRLHQ Q ALPDGAHI ...String: MAVLGLQGVR GGVGTTTITA ALAWSLQMLG ENVLVVDACP DNLLRLSFNV DFTHRQGWAR AMLDGQDWRD AGLRYTSQLD LLPFGQLSI EEQENPQHWQ TRLSDICSGL QQLKASGRYQ WILIDLPRDA SQITHQLLSL CDHSLAIVNV DANCHIRLHQ Q ALPDGAHI LINNFRIGSQ VQDDIYQLWL QSQRRLLPML IHRDEAMAEC LAAKQPVGEY RSDALAAEEI LTLANWCLLN YS GLKTPVG SKS UniProtKB: Cell division protein |
-Macromolecule #3: Protein YhjR
| Macromolecule | Name: Protein YhjR / type: protein_or_peptide / ID: 3 Details: Purified Bcs macrocomplex with stoichiometry BcsA-BcsB6-BcsR2-BcsQ2-BcsE2-BcsF2-BcsG3 Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.645541 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH HHAAGSNNNE PDTLPDPAIG YIFQNDIVAL KQAFSLPDID YADISQREQL AAALKRWPLL AEFAQQK |
-Macromolecule #4: Cellulose biosynthesis protein BcsF
| Macromolecule | Name: Cellulose biosynthesis protein BcsF / type: protein_or_peptide / ID: 4 Details: Purified Bcs macrocomplex with stoichiometry BcsA-BcsB6-BcsR2-BcsQ2-BcsE2-BcsF2-BcsG3 Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.378975 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMTISDIIEI IVVCALIFFP LGYLARHSLR RIRDTLRLFF AKPRYVKPAG TLRRTEKARA TKK |
-Macromolecule #5: 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydr...
| Macromolecule | Name: 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one) type: ligand / ID: 5 / Number of copies: 4 / Formula: C2E |
|---|---|
| Molecular weight | Theoretical: 690.411 Da |
| Chemical component information | 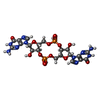 ChemComp-C2E: |
-Macromolecule #6: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 120 mM NaCL 20 mM HEPES pH8 5 mM MgCl2 10 uM ApppCp 4 uM c-di-GMP 0.01% LM-NPG |
| Grid | Model: UltrAuFoil R1.2/1.3 / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
| Details | Purified Bcs macrocomplex with stoichiometry BcsA-BcsB6-BcsR2-BcsQ2-BcsE2-BcsF2-BcsG3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 20022 / Average electron dose: 49.35 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.3 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | |||||||||
| Output model |  PDB-9fp2: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























































