+Search query
-Structure paper
| Title | Tau filaments with the Alzheimer fold in human MAPT mutants V337M and R406W. |
|---|---|
| Journal, issue, pages | Nat Struct Mol Biol, Vol. 32, Issue 7, Page 1297-1304, Year 2025 |
| Publish date | Mar 5, 2025 |
 Authors Authors | Chao Qi / Sofia Lövestam / Alexey G Murzin / Sew Peak-Chew / Catarina Franco / Marika Bogdani / Caitlin Latimer / Jill R Murrell / Patrick W Cullinane / Zane Jaunmuktane / Thomas D Bird / Bernardino Ghetti / Sjors H W Scheres / Michel Goedert /   |
| PubMed Abstract | Frontotemporal dementia (FTD) and Alzheimer's disease (AD) are the most common forms of early-onset dementia. Unlike AD, FTD begins with behavioral changes before the development of cognitive ...Frontotemporal dementia (FTD) and Alzheimer's disease (AD) are the most common forms of early-onset dementia. Unlike AD, FTD begins with behavioral changes before the development of cognitive impairment. Dominantly inherited mutations in MAPT, the microtubule-associated protein tau gene, give rise to cases of FTD and parkinsonism linked to chromosome 17. These individuals develop abundant filamentous tau inclusions in brain cells in the absence of β-amyloid deposits. Here, we used cryo-electron microscopy to determine the structures of tau filaments from the brains of human MAPT mutants V337M and R406W. Both amino acid substitutions gave rise to tau filaments with the Alzheimer fold, which consisted of paired helical filaments in all V337M and R406W cases and of straight filaments in two V337M cases. We also identified another assembly of the Alzheimer fold into triple tau filaments in a V337M case. Filaments assembled from recombinant tau (297-391) with substitution V337M had the Alzheimer fold and showed an increased rate of assembly. |
 External links External links |  Nat Struct Mol Biol / Nat Struct Mol Biol /  PubMed:40044789 / PubMed:40044789 /  PubMed Central PubMed Central |
| Methods | EM (helical sym.) |
| Resolution | 2.3 - 3.3 Å |
| Structure data | EMDB-19846, PDB-9eo7: EMDB-19849: PHF type tau filament from V337M mutant EMDB-19852: PHF type tau filament from V337M mutant EMDB-19854, PDB-9eog: EMDB-19855, PDB-9eoh: |
| Source |
|
 Keywords Keywords | PROTEIN FIBRIL / PHF / tau filament / V337M / TF / R406W |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




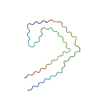



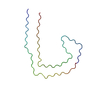




 homo sapiens (human)
homo sapiens (human)