+Search query
-Structure paper
| Title | HflX-mediated drug resistance through ribosome splitting and rRNA disordering in mycobacteria. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 122, Issue 6, Page e2419826122, Year 2025 |
| Publish date | Feb 11, 2025 |
 Authors Authors | Soneya Majumdar / Amuliya Kashyap / Ravi K Koripella / Manjuli R Sharma / Kelley Hurst-Hess / Swati R Manjari / Nilesh K Banavali / Pallavi Ghosh / Rajendra K Agrawal /  |
| PubMed Abstract | HflX is a highly conserved ribosome-associated GTPase implicated in rescuing stalled ribosomes and mediating antibiotic resistance in several bacteria, including macrolide-lincosamide antibiotic ...HflX is a highly conserved ribosome-associated GTPase implicated in rescuing stalled ribosomes and mediating antibiotic resistance in several bacteria, including macrolide-lincosamide antibiotic resistance in mycobacteria. Mycobacterial HflXs carry a distinct N-terminal extension (NTE) and a small insertion, as compared to their eubacterial homologs. Here, we present several high-resolution cryo-EM structures of mycobacterial HflX in complex with the 70S ribosome and its 50S subunit, with and without antibiotics. These structures reveal a distinct mechanism for HflX-mediated ribosome splitting and antibiotic resistance in mycobacteria. Our findings indicate that the NTE of mycobacterial HflX induces persistent disordering of multiple 23S rRNA helices, facilitating the dissociation of the 70S ribosome and generating an inactive pool of 50S subunits. During this process, HflX undergoes a large conformational change that stabilizes its NTE. Mycobacterial HflX also acts as an anti-association factor by binding to predissociated 50S subunits. Our structures show that a mycobacteria-specific insertion in HflX reaches far into the peptidyl transferase center (PTC), such that it would overlap with the ribosome-bound macrolide antibiotics. However, in the presence of antibiotics, this insertion retracts, adjusts around, and interacts with the antibiotic molecules. These results suggest that mycobacterial HflX is agnostic to antibiotic presence in the PTC. It mediates antibiotic resistance by splitting antibiotic-stalled 70S ribosomes and inactivating the resulting 50S subunits. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:39913204 / PubMed:39913204 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.63 - 4.96 Å |
| Structure data | EMDB-43267, PDB-8vio: EMDB-43294, PDB-8vk0: EMDB-43305, PDB-8vk7: EMDB-43317, PDB-8vki: EMDB-43333, PDB-8vkw: EMDB-43409, PDB-8vpk: EMDB-43476, PDB-8vr4: EMDB-43477, PDB-8vr8: EMDB-43484, PDB-8vrl:  EMDB-43778: Structure of HflX mediated, inactive Mycobacterium smegmatis 50S ribosomal subunit  EMDB-43791: Mycobacterium smegmatis 70S ribosome bound to P-tRNA  EMDB-44044: Pre-dissociated Mycobacterium smegmatis 50S ribosomal subunit-HflX-GMPPCP complex |
| Chemicals |  ChemComp-GCP: 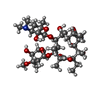 ChemComp-ERY: 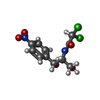 ChemComp-CLM: |
| Source |
|
 Keywords Keywords | RIBOSOME / ribosome splitting / Mycobacterium smegmatis 70S / HflX / TRANSLATION / Mycobacterium smegmatis 50S / disordered 23S rRNA helices / erythromycin / chloramphenicol |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





















 mycolicibacterium smegmatis mc2 155 (bacteria)
mycolicibacterium smegmatis mc2 155 (bacteria)