+Search query
-Structure paper
| Title | Anaerobic cryoEM protocols for air-sensitive nitrogenase proteins. |
|---|---|
| Journal, issue, pages | Nat Protoc, Vol. 19, Issue 7, Page 2026-2051, Year 2024 |
| Publish date | Apr 4, 2024 |
 Authors Authors | Rebeccah A Warmack / Belinda B Wenke / Thomas Spatzal / Douglas C Rees /  |
| PubMed Abstract | Single-particle cryo-electron microscopy (cryoEM) provides an attractive avenue for advancing our atomic resolution understanding of materials, molecules and living systems. However, the vast ...Single-particle cryo-electron microscopy (cryoEM) provides an attractive avenue for advancing our atomic resolution understanding of materials, molecules and living systems. However, the vast majority of published cryoEM methodologies focus on the characterization of aerobically purified samples. Air-sensitive enzymes and microorganisms represent important yet understudied systems in structural biology. We have recently demonstrated the success of an anaerobic single-particle cryoEM workflow applied to the air-sensitive nitrogenase enzymes. In this protocol, we detail the use of Schlenk lines and anaerobic chambers to prepare samples, including a protein tag for monitoring sample exposure to oxygen in air. We describe how to use a plunge freezing apparatus inside of a soft-sided vinyl chamber of the type we routinely use for anaerobic biochemistry and crystallography of oxygen-sensitive proteins. Manual control of the airlock allows for introduction of liquid cryogens into the tent. A custom vacuum port provides slow, continuous evacuation of the tent atmosphere to avoid accumulation of flammable vapors within the enclosed chamber. These methods allowed us to obtain high-resolution structures of both nitrogenase proteins using single-particle cryoEM. The procedures involved can be generally subdivided into a 4 d anaerobic sample generation procedure, and a 1 d anaerobic cryoEM sample preparation step, followed by conventional cryoEM imaging and processing steps. As nitrogen is a substrate for nitrogenase, the Schlenk lines and anaerobic chambers described in this procedure are operated under an argon atmosphere; however, the system and these procedures are compatible with other controlled gas environments. |
 External links External links |  Nat Protoc / Nat Protoc /  PubMed:38575747 PubMed:38575747 |
| Methods | EM (single particle) |
| Resolution | 2.12 - 2.57 Å |
| Structure data | EMDB-27317, PDB-8dby: EMDB-27404, PDB-8dfc: EMDB-27405, PDB-8dfd: EMDB-41151, PDB-8tc3: |
| Chemicals | 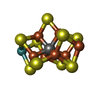 ChemComp-ICS: 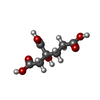 ChemComp-HCA:  ChemComp-FE:  ChemComp-CLF:  ChemComp-HOH:  ChemComp-SF4:  ChemComp-ADP:  ChemComp-ALF:  ChemComp-MG: |
| Source |
|
 Keywords Keywords | METAL BINDING PROTEIN / OXIDOREDUCTASE / Nitrogenase / metalloenzyme / ATPase / metalloprotein / iron-sulfur |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 azotobacter vinelandii (bacteria)
azotobacter vinelandii (bacteria) discosoma sp. lw-2004 (sea anemone)
discosoma sp. lw-2004 (sea anemone)