+Search query
-Structure paper
| Title | X-ray and cryo-EM structures of inhibitor-bound cytochrome complexes for structure-based drug discovery. |
|---|---|
| Journal, issue, pages | IUCrJ, Vol. 5, Issue Pt 2, Page 200-210, Year 2018 |
| Publish date | Mar 1, 2018 |
 Authors Authors | Kangsa Amporndanai / Rachel M Johnson / Paul M O'Neill / Colin W G Fishwick / Alexander H Jamson / Shaun Rawson / Stephen P Muench / S Samar Hasnain / Svetlana V Antonyuk /  |
| PubMed Abstract | Cytochrome , a dimeric multi-subunit electron-transport protein embedded in the inner mitochondrial membrane, is a major drug target for the treatment and prevention of malaria and toxoplasmosis. ...Cytochrome , a dimeric multi-subunit electron-transport protein embedded in the inner mitochondrial membrane, is a major drug target for the treatment and prevention of malaria and toxoplasmosis. Structural studies of cytochrome from mammalian homologues co-crystallized with lead compounds have underpinned structure-based drug design to develop compounds with higher potency and selectivity. However, owing to the limited amount of cytochrome that may be available from parasites, all efforts have been focused on homologous cytochrome complexes from mammalian species, which has resulted in the failure of some drug candidates owing to toxicity in the host. Crystallographic studies of the native parasite proteins are not feasible owing to limited availability of the proteins. Here, it is demonstrated that cytochrome is highly amenable to single-particle cryo-EM (which uses significantly less protein) by solving the apo and two inhibitor-bound structures to ∼4.1 Å resolution, revealing clear inhibitor density at the binding site. Therefore, cryo-EM is proposed as a viable alternative method for structure-based drug discovery using both host and parasite enzymes. |
 External links External links |  IUCrJ / IUCrJ /  PubMed:29765610 / PubMed:29765610 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 3.1 - 4.4 Å |
| Structure data | EMDB-4286, PDB-6fo0: EMDB-4288, PDB-6fo2: EMDB-4292, PDB-6fo6:  PDB-5okd: |
| Chemicals |  ChemComp-PG4:  ChemComp-6PE:  ChemComp-CDL:  ChemComp-HEM: 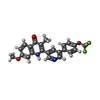 ChemComp-9XE:  ChemComp-LMT:  ChemComp-PEE:  ChemComp-HEC:  ChemComp-PO4:  ChemComp-FES:  ChemComp-PX4:  ChemComp-HOH: 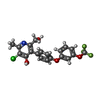 ChemComp-G8U: 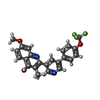 ChemComp-DY2: |
| Source |
|
 Keywords Keywords | ELECTRON TRANSPORT / antimalarial inhibitor / Qi site binder / MEMBRANE PROTEIN / Cryo-EM / Inhibitor binding / cytochrome bc1 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers










