+Search query
-Structure paper
| Title | The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human αβ Integrin via Steric Hindrance. |
|---|---|
| Journal, issue, pages | Structure, Vol. 25, Issue 11, Page 1732-11739.e5, Year 2017 |
| Publish date | Nov 7, 2017 |
 Authors Authors | Andrew J Borst / Zachary M James / William N Zagotta / Mark Ginsberg / Felix A Rey / Frank DiMaio / Marija Backovic / David Veesler /   |
| PubMed Abstract | The LM609 antibody specifically recognizes αβ integrin and inhibits angiogenesis, bone resorption, and viral infections in an arginine-glycine-aspartate-independent manner. LM609 entered phase II ...The LM609 antibody specifically recognizes αβ integrin and inhibits angiogenesis, bone resorption, and viral infections in an arginine-glycine-aspartate-independent manner. LM609 entered phase II clinical trials for the treatment of several cancers and was also used for αβ-targeted radioimmunotherapy. To elucidate the mechanisms of recognition and inhibition of αβ integrin, we solved the structure of the LM609 antigen-binding fragment by X-ray crystallography and determined its binding affinity for αβ. Using single-particle electron microscopy, we show that LM609 binds at the interface between the β-propeller domain of the α chain and the βI domain of the β chain, near the RGD-binding site, of all observed integrin conformational states. Integrating these data with fluorescence size-exclusion chromatography, we demonstrate that LM609 sterically hinders access of large ligands to the RGD-binding pocket, without obstructing it. This work provides a structural framework to expedite future efforts utilizing LM609 as a diagnostic or therapeutic tool. |
 External links External links |  Structure / Structure /  PubMed:29033288 / PubMed:29033288 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.26 - 35.0 Å |
| Structure data | EMDB-7011, PDB-6avq: EMDB-7012, PDB-6avr: EMDB-7013, PDB-6avu:  PDB-5opy: |
| Chemicals |  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | IMMUNE SYSTEM / Antigen-binding fragment / Fab / LM609 / SIGNALING PROTEIN / alpha-V beta-3 integrin / vitaxin / abegrin |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



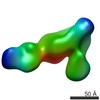

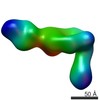



 homo sapiens (human)
homo sapiens (human)
