[English] 日本語
 Yorodumi
Yorodumi- EMDB-7013: Human alpha-V beta-3 Integrin (open conformation) in complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7013 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human alpha-V beta-3 Integrin (open conformation) in complex with the therapeutic antibody LM609 | |||||||||
 Map data Map data | Human alpha-V beta-3 Integrin (open conformation) in complex with the therapeutic antibody LM609 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | alpha-V beta-3 integrin / LM609 / vitaxin / abegrin / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationintegrin alphav-beta8 complex / integrin alphav-beta6 complex / transforming growth factor beta production / negative regulation of entry of bacterium into host cell / integrin alphav-beta5 complex / opsonin binding / integrin alphav-beta1 complex / Cross-presentation of particulate exogenous antigens (phagosomes) / extracellular matrix protein binding / regulation of serotonin uptake ...integrin alphav-beta8 complex / integrin alphav-beta6 complex / transforming growth factor beta production / negative regulation of entry of bacterium into host cell / integrin alphav-beta5 complex / opsonin binding / integrin alphav-beta1 complex / Cross-presentation of particulate exogenous antigens (phagosomes) / extracellular matrix protein binding / regulation of serotonin uptake / positive regulation of adenylate cyclase-inhibiting opioid receptor signaling pathway / tube development / alpha9-beta1 integrin-ADAM8 complex / regulation of trophoblast cell migration / integrin alphaIIb-beta3 complex / regulation of postsynaptic neurotransmitter receptor diffusion trapping / alphav-beta3 integrin-vitronectin complex / maintenance of postsynaptic specialization structure / regulation of extracellular matrix organization / positive regulation of glomerular mesangial cell proliferation / Laminin interactions / platelet alpha granule membrane / integrin alphav-beta3 complex / negative regulation of lipoprotein metabolic process / entry into host cell by a symbiont-containing vacuole / alphav-beta3 integrin-PKCalpha complex / fibrinogen binding / alphav-beta3 integrin-HMGB1 complex / vascular endothelial growth factor receptor 2 binding / negative regulation of lipid transport / regulation of phagocytosis / positive regulation of vascular endothelial growth factor signaling pathway / Elastic fibre formation / mesodermal cell differentiation / cell-substrate junction assembly / alphav-beta3 integrin-IGF-1-IGF1R complex / positive regulation of bone resorption / transforming growth factor beta binding / platelet-derived growth factor receptor binding / glycinergic synapse / positive regulation of small GTPase mediated signal transduction / regulation of release of sequestered calcium ion into cytosol / filopodium membrane / extracellular matrix binding / wound healing, spreading of epidermal cells / positive regulation of vascular endothelial growth factor receptor signaling pathway / apolipoprotein A-I-mediated signaling pathway / positive regulation of cell adhesion mediated by integrin / negative regulation of low-density lipoprotein particle clearance / regulation of bone resorption / angiogenesis involved in wound healing / apoptotic cell clearance / positive regulation of fibroblast migration / integrin complex / positive regulation of smooth muscle cell migration / heterotypic cell-cell adhesion / smooth muscle cell migration / Molecules associated with elastic fibres / cell adhesion mediated by integrin / negative chemotaxis / positive regulation of cell-matrix adhesion / Mechanical load activates signaling by PIEZO1 and integrins in osteocytes / Syndecan interactions / p130Cas linkage to MAPK signaling for integrins / cellular response to insulin-like growth factor stimulus / protein disulfide isomerase activity / regulation of postsynaptic neurotransmitter receptor internalization / positive regulation of osteoblast proliferation / microvillus membrane / cell-substrate adhesion / platelet-derived growth factor receptor signaling pathway / endodermal cell differentiation / PECAM1 interactions / GRB2:SOS provides linkage to MAPK signaling for Integrins / TGF-beta receptor signaling activates SMADs / fibronectin binding / lamellipodium membrane / positive regulation of intracellular signal transduction / blood coagulation, fibrin clot formation / negative regulation of macrophage derived foam cell differentiation / negative regulation of lipid storage / ECM proteoglycans / Integrin cell surface interactions / negative regulation of endothelial cell apoptotic process / positive regulation of T cell migration / vasculogenesis / specific granule membrane / voltage-gated calcium channel activity / coreceptor activity / phagocytic vesicle / cellular response to platelet-derived growth factor stimulus / ERK1 and ERK2 cascade / positive regulation of endothelial cell proliferation / Integrin signaling / extrinsic apoptotic signaling pathway in absence of ligand / positive regulation of substrate adhesion-dependent cell spreading / cell adhesion molecule binding / positive regulation of endothelial cell migration / positive regulation of smooth muscle cell proliferation / substrate adhesion-dependent cell spreading Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 35.0 Å | |||||||||
 Authors Authors | Borst AJ / James ZN | |||||||||
 Citation Citation |  Journal: Structure / Year: 2017 Journal: Structure / Year: 2017Title: The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human αβ Integrin via Steric Hindrance. Authors: Andrew J Borst / Zachary M James / William N Zagotta / Mark Ginsberg / Felix A Rey / Frank DiMaio / Marija Backovic / David Veesler /   Abstract: The LM609 antibody specifically recognizes αβ integrin and inhibits angiogenesis, bone resorption, and viral infections in an arginine-glycine-aspartate-independent manner. LM609 entered phase II ...The LM609 antibody specifically recognizes αβ integrin and inhibits angiogenesis, bone resorption, and viral infections in an arginine-glycine-aspartate-independent manner. LM609 entered phase II clinical trials for the treatment of several cancers and was also used for αβ-targeted radioimmunotherapy. To elucidate the mechanisms of recognition and inhibition of αβ integrin, we solved the structure of the LM609 antigen-binding fragment by X-ray crystallography and determined its binding affinity for αβ. Using single-particle electron microscopy, we show that LM609 binds at the interface between the β-propeller domain of the α chain and the βI domain of the β chain, near the RGD-binding site, of all observed integrin conformational states. Integrating these data with fluorescence size-exclusion chromatography, we demonstrate that LM609 sterically hinders access of large ligands to the RGD-binding pocket, without obstructing it. This work provides a structural framework to expedite future efforts utilizing LM609 as a diagnostic or therapeutic tool. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7013.map.gz emd_7013.map.gz | 2.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7013-v30.xml emd-7013-v30.xml emd-7013.xml emd-7013.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7013.png emd_7013.png | 24.3 KB | ||
| Filedesc metadata |  emd-7013.cif.gz emd-7013.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7013 http://ftp.pdbj.org/pub/emdb/structures/EMD-7013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7013 | HTTPS FTP |
-Related structure data
| Related structure data |  6avuMC  7011C  7012C  5opyC  6avqC  6avrC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7013.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7013.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human alpha-V beta-3 Integrin (open conformation) in complex with the therapeutic antibody LM609 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.2054 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
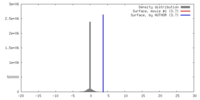
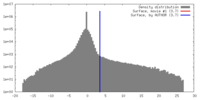
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Quaternary complex of human alpha-V beta-3 integrin with the Fab LM609
| Entire | Name: Quaternary complex of human alpha-V beta-3 integrin with the Fab LM609 |
|---|---|
| Components |
|
-Supramolecule #1: Quaternary complex of human alpha-V beta-3 integrin with the Fab LM609
| Supramolecule | Name: Quaternary complex of human alpha-V beta-3 integrin with the Fab LM609 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 230 KDa |
-Macromolecule #1: Integrin alpha-V
| Macromolecule | Name: Integrin alpha-V / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 105.894188 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FNLDVDSPAE YSGPEGSYFG FAVDFFVPSA SSRMFLLVGA PKANTTQPGI VEGGQVLKCD WSSTRRCQPI EFDATGNRDY AKDDPLEFK SHQWFGASVR SKQDKILACA PLYHWRTEMK QEREPVGTCF LQDGTKTVEY APCRSQDIDA DGQGFCQGGF S IDFTKADR ...String: FNLDVDSPAE YSGPEGSYFG FAVDFFVPSA SSRMFLLVGA PKANTTQPGI VEGGQVLKCD WSSTRRCQPI EFDATGNRDY AKDDPLEFK SHQWFGASVR SKQDKILACA PLYHWRTEMK QEREPVGTCF LQDGTKTVEY APCRSQDIDA DGQGFCQGGF S IDFTKADR VLLGGPGSFY WQGQLISDQV AEIVSKYDPN VYSIKYNNQL ATRTAQAIFD DSYLGYSVAV GDFNGDGIDD FV SGVPRAA RTLGMVYIYD GKNMSSLYNF TGEQMAAYFG FSVAATDING DDYADVFIGA PLFMDRGSDG KLQEVGQVSV SLQ RASGDF QTTKLNGFEV FARFGSAIAP LGDLDQDGFN DIAIAAPYGG EDKKGIVYIF NGRSTGLNAV PSQILEGQWA ARSM PPSFG YSMKGATDID KNGYPDLIVG AFGVDRAILY RARPVITVNA GLEVYPSILN QDNKTCSLPG TALKVSCFNV RFCLK ADGK GVLPRKLNFQ VELLLDKLKQ KGAIRRALFL YSRSPSHSKN MTISRGGLMQ CEELIAYLRD ESEFRDKLTP ITIFME YRL DYRTAADTTG LQPILNQFTP ANISRQAHIL LDCGEDNVCK PKLEVSVDSD QKKIYIGDDN PLTLIVKAQN QGEGAYE AE LIVSIPLQAD FIGVVRNNEA LARLSCAFKT ENQTRQVVCD LGNPMKAGTQ LLAGLRFSVH QQSEMDTSVK FDLQIQSS N LFDKVSPVVS HKVDLAVLAA VEIRGVSSPD HIFLPIPNWE HKENPETEED VGPVVQHIYE LRNNGPSSFS KAMLHLQWP YKYNNNTLLY ILHYDIDGPM NCTSDMEINP LRIKISSLQT TEKNDTVAGQ GERDHLITKR DLALSEGDIH TLGCGVAQCL KIVCQVGRL DRGKSAILYV KSLLWTETFM NKENQNHSYS LKSSASFNVI EFPYKNLPIE DITNSTLVTT NVTWGIQP UniProtKB: Integrin alpha-V |
-Macromolecule #2: Integrin beta-3
| Macromolecule | Name: Integrin beta-3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 76.523125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPNICTTRGV SSCQQCLAVS PMCAWCSDEA LPLGSPRCDL KENLLKDNCA PESIEFPVSE ARVLEDRPLS DKGSGDSSQV TQVSPQRIA LRLRPDDSKN FSIQVRQVED YPVDIYYLMD LSYSMKDDLW SIQNLGTKLA TQMRKLTSNL RIGFGAFVDK P VSPYMYIS ...String: GPNICTTRGV SSCQQCLAVS PMCAWCSDEA LPLGSPRCDL KENLLKDNCA PESIEFPVSE ARVLEDRPLS DKGSGDSSQV TQVSPQRIA LRLRPDDSKN FSIQVRQVED YPVDIYYLMD LSYSMKDDLW SIQNLGTKLA TQMRKLTSNL RIGFGAFVDK P VSPYMYIS PPEALENPCY DMKTTCLPMF GYKHVLTLTD QVTRFNEEVK KQSVSRNRDA PEGGFDAIMQ ATVCDEKIGW RN DASHLLV FTTDAKTHIA LDGRLAGIVQ PNDGQCHVGS DNHYSASTTM DYPSLGLMTE KLSQKNINLI FAVTENVVNL YQN YSELIP GTTVGVLSMD SSNVLQLIVD AYGKIRSKVE LEVRDLPEEL SLSFNATCLN NEVIPGLKSC MGLKIGDTVS FSIE AKVRG CPQEKEKSFT IKPVGFKDSL IVQVTFDCDC ACQAQAEPNS HRCNNGNGTF ECGVCRCGPG WLGSQCECSE EDYRP SQQD ECSPREGQPV CSQRGECLCG QCVCHSSDFG KITGKYCECD DFSCVRYKGE MCSGHGQCSC GDCLCDSDWT GYYCNC TTR TDTCMSSNGL LCSGRGKCEC GSCVCIQPGS YGDTCEKCPT CPDACTFKKE CVECKKFDRG ALHDENTCNR YCRDEIE SV KELKDTGKDA VNCTYKNEDD CVVRFQYYED SSGKSILYVV EEPECPKGPD UniProtKB: Integrin beta-3 |
-Macromolecule #3: Fab LM609 heavy chain
| Macromolecule | Name: Fab LM609 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.2231 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLEESGGG LVKPGGSLKL SCAASGFAFS SYDMSWVRQI PEKRLEWVAK VSSGGGSTYY LDTVQGRFTI SRDNAKNTLY LQMSSLNSE DTAMYYCARH NYGSFAYWGQ GTLVTVSAAK TTPPSVYPLA PGSAAQTNSM VTLGCLVKGY FPEPVTVTWN S GSLSSGVH ...String: EVQLEESGGG LVKPGGSLKL SCAASGFAFS SYDMSWVRQI PEKRLEWVAK VSSGGGSTYY LDTVQGRFTI SRDNAKNTLY LQMSSLNSE DTAMYYCARH NYGSFAYWGQ GTLVTVSAAK TTPPSVYPLA PGSAAQTNSM VTLGCLVKGY FPEPVTVTWN S GSLSSGVH TFPAVLQSDL YTLSSSVTVP SSTWPSETVT CNVAHPASST KVDKKIVPRD CGASDDDDKA GWSHPQFEKG GG SGGGSGG GSWSHPQFEK |
-Macromolecule #4: Fab LM609 light chain
| Macromolecule | Name: Fab LM609 light chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.628977 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ELVMTQTPAT LSVTPGDSVS LSCRASQSIS NHLHWYQQKS HESPRLLIKY ASQSISGIPS RFSGSGSGTD FTLSINSVET EDFGMYFCQ QSNSWPHTFG GGTKLEIKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS ...String: ELVMTQTPAT LSVTPGDSVS LSCRASQSIS NHLHWYQQKS HESPRLLIKY ASQSISGIPS RFSGSGSGTD FTLSINSVET EDFGMYFCQ QSNSWPHTFG GGTKLEIKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS KDSTYSMSST LTLTKDEYER HNSYTCEATH KTSTSPIVKS FNRNEC |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl formate |
| Grid | Model: C-flat 2/0.5 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
- Image processing
Image processing
| Startup model | Type of model: RANDOM CONICAL TILT |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 35.0 Å / Resolution method: FSC 0.5 CUT-OFF / Number images used: 650 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6avu: |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera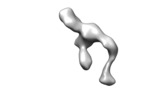

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)