+Search query
-Structure paper
| Title | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation. |
|---|---|
| Journal, issue, pages | Science, Vol. 387, Issue 6733, Page 498-504, Year 2025 |
| Publish date | Jan 31, 2025 |
 Authors Authors | Max Dongsheng Yin / Olivier N Lemaire / José Guadalupe Rosas Jiménez / Mélissa Belhamri / Anna Shevchenko / Gerhard Hummer / Tristan Wagner / Bonnie J Murphy /   |
| PubMed Abstract | In the ancient microbial Wood-Ljungdahl pathway, carbon dioxide (CO) is fixed in a multistep process that ends with acetyl-coenzyme A (acetyl-CoA) synthesis at the bifunctional carbon monoxide ...In the ancient microbial Wood-Ljungdahl pathway, carbon dioxide (CO) is fixed in a multistep process that ends with acetyl-coenzyme A (acetyl-CoA) synthesis at the bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase complex (CODH/ACS). In this work, we present structural snapshots of the CODH/ACS from the gas-converting acetogen , characterizing the molecular choreography of the overall reaction, including electron transfer to the CODH for CO reduction, methyl transfer from the corrinoid iron-sulfur protein (CoFeSP) partner to the ACS active site, and acetyl-CoA production. Unlike CODH, the multidomain ACS undergoes large conformational changes to form an internal connection to the CODH active site, accommodate the CoFeSP for methyl transfer, and protect the reaction intermediates. Altogether, the structures allow us to draw a detailed reaction mechanism of this enzyme, which is crucial for CO fixation in anaerobic organisms. |
 External links External links |  Science / Science /  PubMed:39883773 PubMed:39883773 |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 1.94 - 3.29 Å |
| Structure data | EMDB-50897, PDB-9fzy:  EMDB-50898: Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) from Clostridium autoethanogenum (consensus map)  EMDB-50899: Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with corrinoid iron-sulfur protein (CoFeSP) from Clostridium autoethanogenum (focused refinement, class 3A) EMDB-50900, PDB-9fzz:  EMDB-50901: Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with corrinoid iron-sulfur protein (CoFeSP) from Clostridium autoethanogenum (focused refinement, class 3B) EMDB-50902, PDB-9g00:  EMDB-50903: Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with corrinoid iron-sulfur protein (CoFeSP) from Clostridium autoethanogenum (focused refinement, class 3Cb)  EMDB-50904: Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with corrinoid iron-sulfur protein (CoFeSP) from Clostridium autoethanogenum (focused refinement, class 3Ca) EMDB-50905, PDB-9g01:  EMDB-50906: Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) from Clostridium autoethanogenum (focused refinement, closed and CO-bound state) EMDB-50907, PDB-9g02:  EMDB-50908: Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) from Clostridium autoethanogenum (focused refinement, semi-extended state) EMDB-50909, PDB-9g03:  PDB-9g7i: |
| Chemicals |  ChemComp-SF4:  ChemComp-B12:  ChemComp-NI:  ChemComp-RQM:  ChemComp-CMO:  ChemComp-EDO:  ChemComp-PE4:  ChemComp-GOL:  ChemComp-PEG:  ChemComp-CA:  ChemComp-CL: 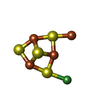 ChemComp-XCC:  ChemComp-ACO:  ChemComp-TRS:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | OXIDOREDUCTASE / anaerobic CO2 fixation / acetyl-CoA synthesis / metalloenzyme / Wood-Ljungdahl pathway. / Carbon monoxide / acetyl-CoA / Wood-Ljungdahl pathway / C-cluster / A-cluster / gas channelling / acetogenic bacteria |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers















 clostridium autoethanogenum dsm 10061 (bacteria)
clostridium autoethanogenum dsm 10061 (bacteria)