[English] 日本語
 Yorodumi
Yorodumi- PDB-9g7i: Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9g7i | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with acetyl-Coenyzme A from Clostridium autoethanogenum | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | OXIDOREDUCTASE / Carbon monoxide / acetyl-CoA / Wood-Ljungdahl pathway / C-cluster / A-cluster / gas channelling / acetogenic bacteria | |||||||||
| Function / homology |  Function and homology information Function and homology informationCO-methylating acetyl-CoA synthase / CO-methylating acetyl-CoA synthase activity / anaerobic carbon-monoxide dehydrogenase activity / acetyl-CoA metabolic process / 4 iron, 4 sulfur cluster binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Clostridium autoethanogenum DSM 10061 (bacteria) Clostridium autoethanogenum DSM 10061 (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.93 Å MOLECULAR REPLACEMENT / Resolution: 2.93 Å | |||||||||
 Authors Authors | Lemaire, O.N. / Yin, M.D. / Murphy, B.J. / Wagner, T. | |||||||||
| Funding support |  Germany, 2items Germany, 2items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2025 Journal: Science / Year: 2025Title: Conformational dynamics of a multienzyme complex in anaerobic carbon fixation. Authors: Max Dongsheng Yin / Olivier N Lemaire / José Guadalupe Rosas Jiménez / Mélissa Belhamri / Anna Shevchenko / Gerhard Hummer / Tristan Wagner / Bonnie J Murphy /   Abstract: In the ancient microbial Wood-Ljungdahl pathway, carbon dioxide (CO) is fixed in a multistep process that ends with acetyl-coenzyme A (acetyl-CoA) synthesis at the bifunctional carbon monoxide ...In the ancient microbial Wood-Ljungdahl pathway, carbon dioxide (CO) is fixed in a multistep process that ends with acetyl-coenzyme A (acetyl-CoA) synthesis at the bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase complex (CODH/ACS). In this work, we present structural snapshots of the CODH/ACS from the gas-converting acetogen , characterizing the molecular choreography of the overall reaction, including electron transfer to the CODH for CO reduction, methyl transfer from the corrinoid iron-sulfur protein (CoFeSP) partner to the ACS active site, and acetyl-CoA production. Unlike CODH, the multidomain ACS undergoes large conformational changes to form an internal connection to the CODH active site, accommodate the CoFeSP for methyl transfer, and protect the reaction intermediates. Altogether, the structures allow us to draw a detailed reaction mechanism of this enzyme, which is crucial for CO fixation in anaerobic organisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9g7i.cif.gz 9g7i.cif.gz | 1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9g7i.ent.gz pdb9g7i.ent.gz | 861.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9g7i.json.gz 9g7i.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g7/9g7i https://data.pdbj.org/pub/pdb/validation_reports/g7/9g7i ftp://data.pdbj.org/pub/pdb/validation_reports/g7/9g7i ftp://data.pdbj.org/pub/pdb/validation_reports/g7/9g7i | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  9fzyC  9fzzC  9g00C  9g01C  9g02C  9g03C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 2 types, 4 molecules ADBC
| #1: Protein | Mass: 76991.070 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)  Clostridium autoethanogenum DSM 10061 (bacteria) Clostridium autoethanogenum DSM 10061 (bacteria)Cell line: / / Organ: / / Plasmid details: / / Variant: wild type / Strain: DSM 10061 / Tissue: / References: UniProt: F8TEQ9, CO-methylating acetyl-CoA synthase #2: Protein | Mass: 67977.531 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)  Clostridium autoethanogenum DSM 10061 (bacteria) Clostridium autoethanogenum DSM 10061 (bacteria)Cell line: / / Organ: / / Plasmid details: / / Variant: wild type / Strain: DSM 10061 / Tissue: / / References: anaerobic carbon monoxide dehydrogenase |
|---|
-Non-polymers , 12 types, 89 molecules 







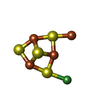














| #3: Chemical | ChemComp-SF4 / #4: Chemical | ChemComp-NI / #5: Chemical | ChemComp-EDO / #6: Chemical | ChemComp-PE4 / | #7: Chemical | ChemComp-GOL / #8: Chemical | #9: Chemical | ChemComp-CA / #10: Chemical | ChemComp-CL / #11: Chemical | #12: Chemical | ChemComp-ACO / | #13: Chemical | ChemComp-TRS / | #14: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.74 Å3/Da / Density % sol: 74.08 % / Description: brown square plate |
|---|---|
| Crystal grow | Temperature: 293.15 K / Method: vapor diffusion, sitting drop / pH: 8.5 Details: Crystallization was performed anaerobically by initial screening at 20 degrees Celsius using the sitting drop method on 96 Well MRC 2 Drop polystyrene Crystallization Plates SWISSCI in a Coy ...Details: Crystallization was performed anaerobically by initial screening at 20 degrees Celsius using the sitting drop method on 96 Well MRC 2 Drop polystyrene Crystallization Plates SWISSCI in a Coy tent containing an N2/H2 (97:3%) atmosphere. The reservoir contained 90 ul of 25 % (w/v) glycerol ethoxylate 1,000, 100 mM Tris/HCl pH 8.5, and 100 mM CaCl2. 0.6 ul of crystallization solution was mixed with 0.6 ul of the protein, containing a final concentration of 2 mM acetyl-CoA. The protein was concentrated at 15 mg/ml in 25 mM Tris/HCl pH 7.6, 10% (v/v) glycerol, and 2 mM dithiothreitol. The crystals appeared in few weeks and were soaked in the crystallization solution supplemented with 15% (v/v) Ethylene glycol for a few seconds before freezing in liquid nitrogen. PH range: / / Temp details: / |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 1.00003 Å / Beamline: X06DA / Wavelength: 1.00003 Å |
| Detector | Type: DECTRIS PILATUS 2M-F / Detector: PIXEL / Date: Jan 29, 2021 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.00003 Å / Relative weight: 1 |
| Reflection | Resolution: 2.93→59.61 Å / Num. obs: 99666 / % possible obs: 96.1 % / Redundancy: 22.1 % / CC1/2: 0.998 / Rmerge(I) obs: 0.207 / Rpim(I) all: 0.045 / Rrim(I) all: 0.211 / Rsym value: 0.045 / Net I/σ(I): 14.5 |
| Reflection shell | Resolution: 2.93→3.16 Å / Redundancy: 23.3 % / Rmerge(I) obs: 2.423 / Mean I/σ(I) obs: 1.6 / Num. unique obs: 4983 / CC1/2: 0.593 / Rpim(I) all: 0.513 / Rrim(I) all: 2.477 / Rsym value: 0.513 / % possible all: 73 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.93→49.68 Å / SU ML: 0.28 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 23.23 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 2.93→49.68 Å / SU ML: 0.28 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 23.23 / Stereochemistry target values: MLDetails: The structure was refined with phenix.refine considering translation libration screw. Riding hydrogens and non-crystallography symmetry were not used during the refinement.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.1 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 89.64 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.93→49.68 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller
















 PDBj
PDBj









