+Search query
-Structure paper
| Title | The structures of protein kinase A in complex with CFTR: Mechanisms of phosphorylation and noncatalytic activation. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 121, Issue 46, Page e2409049121, Year 2024 |
| Publish date | Nov 12, 2024 |
 Authors Authors | Karol Fiedorczuk / Iordan Iordanov / Csaba Mihályi / Andras Szollosi / László Csanády / Jue Chen /   |
| PubMed Abstract | Protein kinase A (PKA) is a key regulator of cellular functions by selectively phosphorylating numerous substrates, including ion channels, enzymes, and transcription factors. It has long served as a ...Protein kinase A (PKA) is a key regulator of cellular functions by selectively phosphorylating numerous substrates, including ion channels, enzymes, and transcription factors. It has long served as a model system for understanding the eukaryotic kinases. Using cryoelectron microscopy, we present complex structures of the PKA catalytic subunit (PKA-C) bound to a full-length protein substrate, the cystic fibrosis transmembrane conductance regulator (CFTR)-an ion channel vital to human health. CFTR gating requires phosphorylation of its regulatory (R) domain. Unphosphorylated CFTR engages PKA-C at two locations, establishing two "catalytic stations" near to, but not directly involving, the R domain. This configuration, coupled with the conformational flexibility of the R domain, permits transient interactions of the eleven spatially separated phosphorylation sites. Furthermore, we determined two structures of the open-pore CFTR stabilized by PKA-C, providing a molecular basis to understand how PKA-C stimulates CFTR currents even in the absence of phosphorylation. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:39495916 / PubMed:39495916 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.8 - 9.0 Å |
| Structure data | EMDB-47235, PDB-9dw4: EMDB-47236, PDB-9dw5: EMDB-47237, PDB-9dw7: EMDB-47238, PDB-9dw8: EMDB-47239, PDB-9dw9: |
| Chemicals |  ChemComp-MG:  ChemComp-ANP: 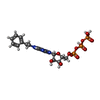 ChemComp-B44:  ChemComp-CLR:  ChemComp-D10:  ChemComp-ATP:  ChemComp-CL: 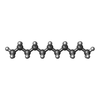 ChemComp-UND:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | HYDROLASE / CFTR / PKA / complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers














 homo sapiens (human)
homo sapiens (human)