+Search query
-Structure paper
| Title | Structural basis for human Ca1.2 inhibition by multiple drugs and the neurotoxin calciseptine. |
|---|---|
| Journal, issue, pages | Cell, Vol. 186, Issue 24, Page 5363-5374.e16, Year 2023 |
| Publish date | Nov 22, 2023 |
 Authors Authors | Shuai Gao / Xia Yao / Jiaofeng Chen / Gaoxingyu Huang / Xiao Fan / Lingfeng Xue / Zhangqiang Li / Tong Wu / Yupeng Zheng / Jian Huang / Xueqin Jin / Yan Wang / Zhifei Wang / Yong Yu / Lei Liu / Xiaojing Pan / Chen Song / Nieng Yan /   |
| PubMed Abstract | Ca1.2 channels play crucial roles in various neuronal and physiological processes. Here, we present cryo-EM structures of human Ca1.2, both in its apo form and in complex with several drugs, as well ...Ca1.2 channels play crucial roles in various neuronal and physiological processes. Here, we present cryo-EM structures of human Ca1.2, both in its apo form and in complex with several drugs, as well as the peptide neurotoxin calciseptine. Most structures, apo or bound to calciseptine, amlodipine, or a combination of amiodarone and sofosbuvir, exhibit a consistent inactivated conformation with a sealed gate, three up voltage-sensing domains (VSDs), and a down VSD. Calciseptine sits on the shoulder of the pore domain, away from the permeation path. In contrast, when pinaverium bromide, an antispasmodic drug, is inserted into a cavity reminiscent of the IFM-binding site in Na channels, a series of structural changes occur, including upward movement of VSD coupled with dilation of the selectivity filter and its surrounding segments in repeat III. Meanwhile, S4-5 merges with S5 to become a single helix, resulting in a widened but still non-conductive intracellular gate. |
 External links External links |  Cell / Cell /  PubMed:37972591 PubMed:37972591 |
| Methods | EM (single particle) |
| Resolution | 2.9 - 3.3 Å |
| Structure data | EMDB-29102, PDB-8fhs: EMDB-37472, PDB-8we6: EMDB-37473, PDB-8we7: EMDB-37474, PDB-8we8: EMDB-37475, PDB-8we9: EMDB-37476, PDB-8wea: |
| Chemicals |  ChemComp-CA: 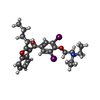 ChemComp-BBI: 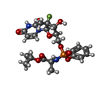 ChemComp-WG6:  ChemComp-3PE:  ChemComp-CLR:  ChemComp-NAG:  ChemComp-PT5: 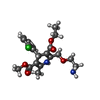 ChemComp-6UB:  ChemComp-9Z9: 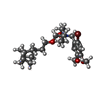 ChemComp-WB9: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN / Cav1.2 / Channels / Calcium Ion-Selective / MEMBRANE PROTEIN |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers















 homo sapiens (human)
homo sapiens (human) dendroaspis polylepis polylepis (black mamba)
dendroaspis polylepis polylepis (black mamba)