[English] 日本語
 Yorodumi
Yorodumi- EMDB-37472: Human L-type voltage-gated calcium channel Cav1.2 at 2.9 Angstrom... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human L-type voltage-gated calcium channel Cav1.2 at 2.9 Angstrom resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cav1.2 / Channels / Calcium Ion-Selective / TRANSPORT PROTEIN / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of high voltage-gated calcium channel activity / voltage-gated calcium channel activity involved in AV node cell action potential / voltage-gated calcium channel activity involved in cardiac muscle cell action potential / immune system development / positive regulation of adenylate cyclase activity / Presynaptic depolarization and calcium channel opening / regulation of membrane repolarization during action potential / membrane depolarization during atrial cardiac muscle cell action potential / calcium ion transmembrane transport via high voltage-gated calcium channel / Phase 2 - plateau phase ...positive regulation of high voltage-gated calcium channel activity / voltage-gated calcium channel activity involved in AV node cell action potential / voltage-gated calcium channel activity involved in cardiac muscle cell action potential / immune system development / positive regulation of adenylate cyclase activity / Presynaptic depolarization and calcium channel opening / regulation of membrane repolarization during action potential / membrane depolarization during atrial cardiac muscle cell action potential / calcium ion transmembrane transport via high voltage-gated calcium channel / Phase 2 - plateau phase / high voltage-gated calcium channel activity / membrane depolarization during AV node cell action potential / membrane depolarization during bundle of His cell action potential / cardiac conduction / L-type voltage-gated calcium channel complex / membrane depolarization during cardiac muscle cell action potential / positive regulation of muscle contraction / cell communication by electrical coupling involved in cardiac conduction / regulation of ventricular cardiac muscle cell action potential / NCAM1 interactions / regulation of ventricular cardiac muscle cell membrane repolarization / camera-type eye development / cardiac muscle cell action potential involved in contraction / embryonic forelimb morphogenesis / calcium ion transport into cytosol / regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / voltage-gated calcium channel complex / Mechanical load activates signaling by PIEZO1 and integrins in osteocytes / Phase 0 - rapid depolarisation / alpha-actinin binding / regulation of heart rate by cardiac conduction / calcium ion import across plasma membrane / regulation of calcium ion transport / neuronal dense core vesicle / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / voltage-gated calcium channel activity / presynaptic active zone membrane / sarcoplasmic reticulum / protein localization to plasma membrane / calcium channel regulator activity / Regulation of insulin secretion / postsynaptic density membrane / calcium ion transmembrane transport / GABA-ergic synapse / Z disc / cellular response to amyloid-beta / Adrenaline,noradrenaline inhibits insulin secretion / calcium ion transport / T cell receptor signaling pathway / heart development / positive regulation of cytosolic calcium ion concentration / perikaryon / chemical synaptic transmission / calmodulin binding / postsynaptic density / cilium / synapse / dendrite / extracellular exosome / nucleoplasm / metal ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Gao S / Yao X / Yan N | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Structural basis for human Ca1.2 inhibition by multiple drugs and the neurotoxin calciseptine. Authors: Shuai Gao / Xia Yao / Jiaofeng Chen / Gaoxingyu Huang / Xiao Fan / Lingfeng Xue / Zhangqiang Li / Tong Wu / Yupeng Zheng / Jian Huang / Xueqin Jin / Yan Wang / Zhifei Wang / Yong Yu / Lei ...Authors: Shuai Gao / Xia Yao / Jiaofeng Chen / Gaoxingyu Huang / Xiao Fan / Lingfeng Xue / Zhangqiang Li / Tong Wu / Yupeng Zheng / Jian Huang / Xueqin Jin / Yan Wang / Zhifei Wang / Yong Yu / Lei Liu / Xiaojing Pan / Chen Song / Nieng Yan /   Abstract: Ca1.2 channels play crucial roles in various neuronal and physiological processes. Here, we present cryo-EM structures of human Ca1.2, both in its apo form and in complex with several drugs, as well ...Ca1.2 channels play crucial roles in various neuronal and physiological processes. Here, we present cryo-EM structures of human Ca1.2, both in its apo form and in complex with several drugs, as well as the peptide neurotoxin calciseptine. Most structures, apo or bound to calciseptine, amlodipine, or a combination of amiodarone and sofosbuvir, exhibit a consistent inactivated conformation with a sealed gate, three up voltage-sensing domains (VSDs), and a down VSD. Calciseptine sits on the shoulder of the pore domain, away from the permeation path. In contrast, when pinaverium bromide, an antispasmodic drug, is inserted into a cavity reminiscent of the IFM-binding site in Na channels, a series of structural changes occur, including upward movement of VSD coupled with dilation of the selectivity filter and its surrounding segments in repeat III. Meanwhile, S4-5 merges with S5 to become a single helix, resulting in a widened but still non-conductive intracellular gate. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37472.map.gz emd_37472.map.gz | 79.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37472-v30.xml emd-37472-v30.xml emd-37472.xml emd-37472.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37472.png emd_37472.png | 37.4 KB | ||
| Filedesc metadata |  emd-37472.cif.gz emd-37472.cif.gz | 9 KB | ||
| Others |  emd_37472_half_map_1.map.gz emd_37472_half_map_1.map.gz emd_37472_half_map_2.map.gz emd_37472_half_map_2.map.gz | 77.6 MB 77.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37472 http://ftp.pdbj.org/pub/emdb/structures/EMD-37472 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37472 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37472 | HTTPS FTP |
-Related structure data
| Related structure data |  8we6MC  8fhsC  8we7C  8we8C  8we9C  8weaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37472.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37472.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.114 Å | ||||||||||||||||||||||||||||||||||||
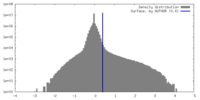
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37472_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
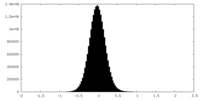
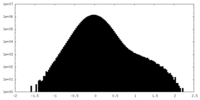
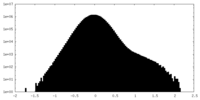
| Density Histograms |
-Half map: #1
| File | emd_37472_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
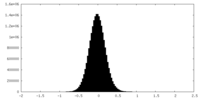
| Density Histograms |
- Sample components
Sample components
-Entire : Cav1.2
| Entire | Name: Cav1.2 |
|---|---|
| Components |
|
-Supramolecule #1: Cav1.2
| Supramolecule | Name: Cav1.2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Voltage-dependent L-type calcium channel subunit alpha-1C
| Macromolecule | Name: Voltage-dependent L-type calcium channel subunit alpha-1C type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 246.85125 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVNENTRMYI PEENHQGSNY GSPRPAHANM NANAAAGLAP EHIPTPGAAL SWQAAIDAAR QAKLMGSAGN ATISTVSSTQ RKRQQYGKP KKQGSTTATR PPRALLCLTL KNPIRRACIS IVEWKPFEII ILLTIFANCV ALAIYIPFPE DDSNATNSNL E RVEYLFLI ...String: MVNENTRMYI PEENHQGSNY GSPRPAHANM NANAAAGLAP EHIPTPGAAL SWQAAIDAAR QAKLMGSAGN ATISTVSSTQ RKRQQYGKP KKQGSTTATR PPRALLCLTL KNPIRRACIS IVEWKPFEII ILLTIFANCV ALAIYIPFPE DDSNATNSNL E RVEYLFLI IFTVEAFLKV IAYGLLFHPN AYLRNGWNLL DFIIVVVGLF SAILEQATKA DGANALGGKG AGFDVKALRA FR VLRPLRL VSGVPSLQVV LNSIIKAMVP LLHIALLVLF VIIIYAIIGL ELFMGKMHKT CYNQEGIADV PAEDDPSPCA LET GHGRQC QNGTVCKPGW DGPKHGITNF DNFAFAMLTV FQCITMEGWT DVLYWVNDAV GRDWPWIYFV TLIIIGSFFV LNLV LGVLS GEFSKEREKA KARGDFQKLR EKQQLEEDLK GYLDWITQAE DIDPENEDEG MDEEKPRNMS MPTSETESVN TENVA GGDI EGENCGARLA HRISKSKFSR YWRRWNRFCR RKCRAAVKSN VFYWLVIFLV FLNTLTIASE HYNQPNWLTE VQDTAN KAL LALFTAEMLL KMYSLGLQAY FVSLFNRFDC FVVCGGILET ILVETKIMSP LGISVLRCVR LLRIFKITRY WNSLSNL VA SLLNSVRSIA SLLLLLFLFI IIFSLLGMQL FGGKFNFDEM QTRRSTFDNF PQSLLTVFQI LTGEDWNSVM YDGIMAYG G PSFPGMLVCI YFIILFICGN YILLNVFLAI AVDNLADAES LTSAQKEEEE EKERKKLART ASPEKKQELV EKPAVGESK EEKIELKSIT ADGESPPATK INMDDLQPNE NEDKSPYPNP ETTGEEDEEE PEMPVGPRPR PLSELHLKEK AVPMPEASAF FIFSSNNRF RLQCHRIVND TIFTNLILFF ILLSSISLAA EDPVQHTSFR NHILGNADYV FTSIFTLEII LKMTAYGAFL H KGSFCRNY FNILDLLVVS VSLISFGIQS SAINVVKILR VLRVLRPLRA INRAKGLKHV VQCVFVAIRT IGNIVIVTTL LQ FMFACIG VQLFKGKLYT CSDSSKQTEA ECKGNYITYK DGEVDHPIIQ PRSWENSKFD FDNVLAAMMA LFTVSTFEGW PEL LYRSID SHTEDKGPIY NYRVEISIFF IIYIIIIAFF MMNIFVGFVI VTFQEQGEQE YKNCELDKNQ RQCVEYALKA RPLR RYIPK NQHQYKVWYV VNSTYFEYLM FVLILLNTIC LAMQHYGQSC LFKIAMNILN MLFTGLFTVE MILKLIAFKP KGYFS DPWN VFDFLIVIGS IIDVILSETN HYFCDAWNTF DALIVVGSIV DIAITEVNPA EHTQCSPSMN AEENSRISIT FFRLFR VMR LVKLLSRGEG IRTLLWTFIK SFQALPYVAL LIVMLFFIYA VIGMQVFGKI ALNDTTEINR NNNFQTFPQA VLLLFRC AT GEAWQDIMLA CMPGKKCAPE SEPSNSTEGE TPCGSSFAVF YFISFYMLCA FLIINLFVAV IMDNFDYLTR DWSILGPH H LDEFKRIWAE YDPEAKGRIK HLDVVTLLRR IQPPLGFGKL CPHRVACKRL VSMNMPLNSD GTVMFNATLF ALVRTALRI KTEGNLEQAN EELRAIIKKI WKRTSMKLLD QVVPPAGDDE VTVGKFYATF LIQEYFRKFK KRKEQGLVGK PSQRNALSLQ AGLRTLHDI GPEIRRAISG DLTAEEELDK AMKEAVSAAS EDDIFRRAGG LFGNHVSYYQ SDGRSAFPQT FTTQRPLHIN K AGSSQGDT ESPSHEKLVD STFTPSSYSS TGSNANINNA NNTALGRLPR PAGYPSTVST VEGHGPPLSP AIRVQEVAWK LS SNRERHV PMCEDLELRR DSGSAGTQAH CLLLRKANPS RCHSRESQAA MAGQEETSQD ETYEVKMNHD TEACSEPSLL STE MLSYQD DENRQLTLPE EDKRDIRQSP KRGFLRSASL GRRASFHLEC LKRQKDRGGD ISQKTVLPLH LVHHQALAVA GLSP LLQRS HSPASFPRPF ATPPATPGSR GWPPQPVPTL RLEGVESSEK LNSSFPSIHC GSWAETTPGG GGSSAARRVR PVSLM VPSQ AGAPGRQFHG SASSLVEAVL ISEGLGQFAQ DPKFIEVTTQ ELADACDMTI EEMESAADNI LSGGAPQSPN GALLPF VNC RDAGQDRAGG EEDAGCVRAR GRPSEEELQD SRVYVSSL UniProtKB: Voltage-dependent L-type calcium channel subunit alpha-1C |
-Macromolecule #2: Voltage-dependent calcium channel subunit alpha-2/delta-1
| Macromolecule | Name: Voltage-dependent calcium channel subunit alpha-2/delta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 124.692469 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAGCLLALT LTLFQSLLIG PSSEEPFPSA VTIKSWVDKM QEDLVTLAKT ASGVNQLVDI YEKYQDLYTV EPNNARQLVE IAARDIEKL LSNRSKALVR LALEAEKVQA AHQWREDFAS NEVVYYNAKD DLDPEKNDSE PGSQRIKPVF IEDANFGRQI S YQHAAVHI ...String: MAAGCLLALT LTLFQSLLIG PSSEEPFPSA VTIKSWVDKM QEDLVTLAKT ASGVNQLVDI YEKYQDLYTV EPNNARQLVE IAARDIEKL LSNRSKALVR LALEAEKVQA AHQWREDFAS NEVVYYNAKD DLDPEKNDSE PGSQRIKPVF IEDANFGRQI S YQHAAVHI PTDIYEGSTI VLNELNWTSA LDEVFKKNRE EDPSLLWQVF GSATGLARYY PASPWVDNSR TPNKIDLYDV RR RPWYIQG AASPKDMLIL VDVSGSVSGL TLKLIRTSVS EMLETLSDDD FVNVASFNSN AQDVSCFQHL VQANVRNKKV LKD AVNNIT AKGITDYKKG FSFAFEQLLN YNVSRANCNK IIMLFTDGGE ERAQEIFNKY NKDKKVRVFT FSVGQHNYDR GPIQ WMACE NKGYYYEIPS IGAIRINTQE YLDVLGRPMV LAGDKAKQVQ WTNVYLDALE LGLVITGTLP VFNITGQFEN KTNLK NQLI LGVMGVDVSL EDIKRLTPRF TLCPNGYYFA IDPNGYVLLH PNLQPKPIGV GIPTINLRKR RPNIQNPKSQ EPVTLD FLD AELENDIKVE IRNKMIDGES GEKTFRTLVK SQDERYIDKG NRTYTWTPVN GTDYSLALVL PTYSFYYIKA KLEETIT QA RYSETLKPDN FEESGYTFIA PRDYCNDLKI SDNNTEFLLN FNEFIDRKTP NNPSCNADLI NRVLLDAGFT NELVQNYW S KQKNIKGVKA RFVVTDGGIT RVYPKEAGEN WQENPETYED SFYKRSLDND NYVFTAPYFN KSGPGAYESG IMVSKAVEI YIQGKLLKPA VVGIKIDVNS WIENFTKTSI RDPCAGPVCD CKRNSDVMDC VILDDGGFLL MANHDDYTNQ IGRFFGEIDP SLMRHLVNI SVYAFNKSYD YQSVCEPGAA PKQGAGHRSA YVPSVADILQ IGWWATAAAW SILQQFLLSL TFPRLLEAVE M EDDDFTAS LSKQSCITEQ TQYFFDNDSK SFSGVLDCGN CSRIFHGEKL MNTNLIFIMV ESKGTCPCDT RLLIQAEQTS DG PNPCDMV KQPRYRKGPD VCFDNNVLED YTDCGGVSGL NPSLWYIIGI QFLLLWLVSG STHRLL UniProtKB: Voltage-dependent calcium channel subunit alpha-2/delta-1 |
-Macromolecule #3: Voltage-dependent L-type calcium channel subunit beta-3
| Macromolecule | Name: Voltage-dependent L-type calcium channel subunit beta-3 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.607852 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MYDDSYVPGF EDSEAGSADS YTSRPSLDSD VSLEEDRESA RREVESQAQQ QLERAKHKPV AFAVRTNVSY CGVLDEECPV QGSGVNFEA KDFLHIKEKY SNDWWIGRLV KEGGDIAFIP SPQRLESIRL KQEQKARRSG NPSSLSDIGN RRSPPPSLAK Q KQKQAEHV ...String: MYDDSYVPGF EDSEAGSADS YTSRPSLDSD VSLEEDRESA RREVESQAQQ QLERAKHKPV AFAVRTNVSY CGVLDEECPV QGSGVNFEA KDFLHIKEKY SNDWWIGRLV KEGGDIAFIP SPQRLESIRL KQEQKARRSG NPSSLSDIGN RRSPPPSLAK Q KQKQAEHV PPYDVVPSMR PVVLVGPSLK GYEVTDMMQK ALFDFLKHRF DGRISITRVT ADLSLAKRSV LNNPGKRTII ER SSARSSI AEVQSEIERI FELAKSLQLV VLDADTINHP AQLAKTSLAP IIVFVKVSSP KVLQRLIRSR GKSQMKHLTV QMM AYDKLV QCPPESFDVI LDENQLEDAC EHLAEYLEVY WRATHHPAPG PGLLGPPSAI PGLQNQQLLG ERGEEHSPLE RDSL MPSDE ASESSRQAWT GSSQRSSRHL EEDYADAYQD LYQPHRQHTS GLPSANGHDP QDRLLAQDSE HNHSDRNWQR NRPWP KDSY UniProtKB: Voltage-dependent L-type calcium channel subunit beta-3 |
-Macromolecule #7: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #8: 1,2-Distearoyl-sn-glycerophosphoethanolamine
| Macromolecule | Name: 1,2-Distearoyl-sn-glycerophosphoethanolamine / type: ligand / ID: 8 / Number of copies: 1 / Formula: 3PE |
|---|---|
| Molecular weight | Theoretical: 748.065 Da |
| Chemical component information |  ChemComp-3PE: |
-Macromolecule #9: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 9 / Number of copies: 4 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #10: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 10 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #11: [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tr...
| Macromolecule | Name: [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate type: ligand / ID: 11 / Number of copies: 1 / Formula: PT5 |
|---|---|
| Molecular weight | Theoretical: 1.047088 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.9000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)