-Search query
-Search result
Showing 1 - 50 of 56 items for (author: upadhyay & v)

EMDB-28092: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-093

EMDB-28090: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-040

EMDB-28091: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-045
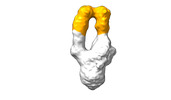
EMDB-28093: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-156
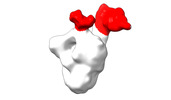
EMDB-28094: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-234

EMDB-28095: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-260

EMDB-28096: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-279

EMDB-28097: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-290
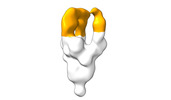
EMDB-28098: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-294

EMDB-28099: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-295

EMDB-28100: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-299

EMDB-28102: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-334
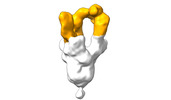
EMDB-28103: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-360

EMDB-28104: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-361

EMDB-28105: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-362

EMDB-28106: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-368

EMDB-28168: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-292

EMDB-28169: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-333
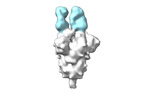
EMDB-28170: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-355

EMDB-28171: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-371

EMDB-23778: 
Negative stain EM map of BG505 SOSIP in complex with Rh4O9.8 monoclonal antibody (as Fab fragment)

EMDB-23779: 
BG505 SOSIP.v5.2 in complex with the polyclonal Fab pAbC-1 from animal Rh.4O9

EMDB-23780: 
BG505 SOSIP MD39 in complex with the monoclonal antibodies Rh.33104 mAb.1 and RM20A3

EMDB-23801: 
BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibodies Rh.33172 mAb.1 and RM19R

PDB-7mdt: 
BG505 SOSIP.v5.2 in complex with the monoclonal antibody Rh4O9.8 (as Fab fragment)

PDB-7mdu: 
BG505 SOSIP MD39 in complex with the monoclonal antibodies Rh.33104 mAb.1 and RM20A3

PDB-7mep: 
BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibodies Rh.33172 mAb.1 and RM19R
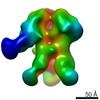
EMDB-9175: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate 224-13
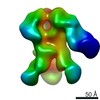
EMDB-9176: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RCn
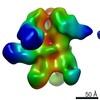
EMDB-9177: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RFr16

EMDB-9178: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate ROw16

EMDB-9179: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RQq16

EMDB-9180: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RRk16
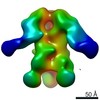
EMDB-9181: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RVh16
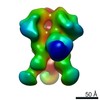
EMDB-9182: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RTh16
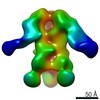
EMDB-9183: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RWh16
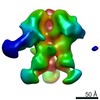
EMDB-9184: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RWr16
EMDB-9185: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RYm16
EMDB-9186: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate ROw16
EMDB-0569: 
HIV-1 vaccine elicited polyclonal 5N6 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
EMDB-0570: 
HIV-1 vaccine elicited polyclonal 99-13 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2

EMDB-0571: 
HIV-1 vaccine elicited polyclonal 145-11 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
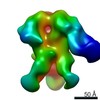
EMDB-0572: 
HIV-1 vaccine elicited polyclonal BM57 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2

EMDB-0573: 
HIV-1 vaccine elicited polyclonal BO77 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2

EMDB-0574: 
HIV-1 vaccine elicited polyclonal BO77 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
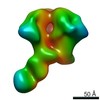
EMDB-0575: 
HIV-1 vaccine elicited polyclonal RAa14 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2

EMDB-0576: 
HIV-1 vaccine elicited polyclonal RAv16 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2

EMDB-0577: 
HIV-1 vaccine elicited polyclonal RAv16 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
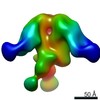
EMDB-0578: 
HIV-1 vaccine elicited polyclonal REj15 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2

EMDB-0579: 
HIV-1 vaccine elicited polyclonal REv16 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
