[English] 日本語
 Yorodumi
Yorodumi- EMDB-23778: Negative stain EM map of BG505 SOSIP in complex with Rh4O9.8 mono... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23778 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain EM map of BG505 SOSIP in complex with Rh4O9.8 monoclonal antibody (as Fab fragment) | |||||||||
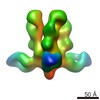
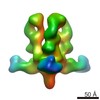
 Map data Map data | 3D map of BG505 SOSIP in complex with the monoclonal antibody Rh4O9.8 (as Fab fragment). Obtained by negative stain EM. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus Human immunodeficiency virus | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 15.9 Å | |||||||||
 Authors Authors | Antanasijevic A / Ward AB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: From structure to sequence: Antibody discovery using cryoEM. Authors: Aleksandar Antanasijevic / Charles A Bowman / Robert N Kirchdoerfer / Christopher A Cottrell / Gabriel Ozorowski / Amit A Upadhyay / Kimberly M Cirelli / Diane G Carnathan / Chiamaka A ...Authors: Aleksandar Antanasijevic / Charles A Bowman / Robert N Kirchdoerfer / Christopher A Cottrell / Gabriel Ozorowski / Amit A Upadhyay / Kimberly M Cirelli / Diane G Carnathan / Chiamaka A Enemuo / Leigh M Sewall / Bartek Nogal / Fangzhu Zhao / Bettina Groschel / William R Schief / Devin Sok / Guido Silvestri / Shane Crotty / Steven E Bosinger / Andrew B Ward /  Abstract: One of the rate-limiting steps in analyzing immune responses to vaccines or infections is the isolation and characterization of monoclonal antibodies. Here, we present a hybrid structural and ...One of the rate-limiting steps in analyzing immune responses to vaccines or infections is the isolation and characterization of monoclonal antibodies. Here, we present a hybrid structural and bioinformatic approach to directly assign the heavy and light chains, identify complementarity-determining regions, and discover sequences from cryoEM density maps of serum-derived polyclonal antibodies bound to an antigen. When combined with next-generation sequencing of immune repertoires, we were able to specifically identify clonal family members, synthesize the monoclonal antibodies, and confirm that they interact with the antigen in a manner equivalent to the corresponding polyclonal antibodies. This structure-based approach for identification of monoclonal antibodies from polyclonal sera opens new avenues for analysis of immune responses and iterative vaccine design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23778.map.gz emd_23778.map.gz | 32.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23778-v30.xml emd-23778-v30.xml emd-23778.xml emd-23778.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23778.png emd_23778.png | 50.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23778 http://ftp.pdbj.org/pub/emdb/structures/EMD-23778 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23778 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23778 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23778.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23778.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map of BG505 SOSIP in complex with the monoclonal antibody Rh4O9.8 (as Fab fragment). Obtained by negative stain EM. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.77 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : BG505 SOSIP.v5.2 in complex with the monoclonal antibody Rh4O9.8 ...
| Entire | Name: BG505 SOSIP.v5.2 in complex with the monoclonal antibody Rh4O9.8 (as Fab fragment) |
|---|---|
| Components |
|
-Supramolecule #1: BG505 SOSIP.v5.2 in complex with the monoclonal antibody Rh4O9.8 ...
| Supramolecule | Name: BG505 SOSIP.v5.2 in complex with the monoclonal antibody Rh4O9.8 (as Fab fragment) type: complex / ID: 1 / Parent: 0 Details: Fab fragment was generated by proteolytic cleavage of IgG antibody |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus Human immunodeficiency virus |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F / Recombinant plasmid: pPPI4 Homo sapiens (human) / Recombinant cell: HEK293F / Recombinant plasmid: pPPI4 |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: TBS, 0.2um filtered | |||||||||
| Staining | Type: NEGATIVE / Material: Uranyl formate / Details: The samples were stained with UF for 60s. | |||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Details | The complex was generated by incubation of Rh4O9.8 monoclonal antibody (as Fab) with recombinantly expressed BG505 SOSIP and subsequent SEC purification. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 62000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















