-Search query
-Search result
Showing all 38 items for (author: chen & xm)

EMDB-63995: 
The structure of mCAT1 in complex with its substrate ornithine and the RBD of FrMLV.
Method: single particle / : Xia LY, Yang Y, Chen XM

EMDB-17766: 
CryoEM structure of Nal1 protein, allele SPIKE, from Oryza sativa japonica group
Method: single particle / : Huang LY, Rety S, Xi XG

EMDB-17768: 
CryoEM structure of Nal1 protein, allele IR64, from Oryza sativa indica cultivar
Method: single particle / : Huang LY, Rety S, Xi XG

EMDB-37849: 
Cryo-EM structure of CB1-beta-arrestin1 complex
Method: single particle / : Liao Y, Zhang H, Shen Q, Cai C

EMDB-35297: 
Structural insights into human brain gut peptide cholecystokinin receptors
Method: single particle / : Ding Y, Zhang H, Liao Y, Chen L, Ji S

EMDB-34259: 
cryo-EM structure of Omicron BA.5 S protein in complex with XGv282
Method: single particle / : Xia XY, Zhang YY, Chi XM, Huang BD, Wu LS, Zhou Q

EMDB-34261: 
cryo-EM structure of Omicron BA.5 S protein in complex with XGv289
Method: single particle / : Xia XY, Zhang YY, Chi XM, Huang BD, Wu LS, Zhou Q

EMDB-34263: 
cryo-EM structure of Omicron BA.5 S protein in complex with S2L20
Method: single particle / : Xia XY, Zhang YY, Chi XM, Huang BD, Wu LS, Zhou Q

EMDB-34023: 
EM structure of human PA28gamma
Method: single particle / : Chen DD, Hao J, Shen CH, Yun CH

EMDB-34024: 
EM structure of human PA28gamma (wild-type)
Method: single particle / : Chen DD, Hao J, Yun CH

EMDB-33359: 
Structural insights into human brain gut peptide cholecystokinin receptors
Method: single particle / : Ding Y, Zhang H, Liao Y, Chen L, Ji S

EMDB-33360: 
Structural insights into human brain gut peptide cholecystokinin receptors
Method: single particle / : Ding Y, Zhang H, Liao Y, Chen L, Ji S

EMDB-33361: 
Structural insights into human brain gut peptide cholecystokinin receptors
Method: single particle / : Ding Y, Zhang H, Liao Y, Chen L, Ji S

EMDB-30049: 
cryo EM map of KCC3
Method: single particle / : Chi XM, Li XR

EMDB-30058: 
KCC3 bound with DIOA
Method: single particle / : Chi XM, Li XR

EMDB-30061: 
cryo_EM map of KCC2
Method: single particle / : Chi XM, Li XR

EMDB-30050: 
Molecular basis for regulation of human potassium chloride cotransporters
Method: single particle / : Chi XM, Li XR, Chen Y, Zhang YY, Su Q, Zhou Q

EMDB-30051: 
cryo_EM map of KCC3, focused refined on transmembrane region
Method: single particle / : Chi XM, Li XR, Chen Y, Zhang YY, Su Q, Zhou Q

EMDB-30059: 
cryo_EM map of KCC3 bound with DIOA, focused refined on extracellular region
Method: single particle / : Chi XM, Li XR, Chen Y, Zhang YY, Su Q, Zhou Q

EMDB-30060: 
cryo_EM map of KCC3 bound with DIOA, focused refined on transmembrane region
Method: single particle / : Chi XM, Li XR, Chen Y, Zhang YY, Su Q, Zhou Q

EMDB-30062: 
cryo_EM map of KCC2, focused refined on extracellular region
Method: single particle / : Chi XM, Li XR, Chen Y, Zhang YY, Su Q, Zhou Q

EMDB-30063: 
cryo_EM map of KCC2, focused refined on transmembrane region
Method: single particle / : Chi XM, Li XR, Chen Y, Zhang YY, Su Q, Zhou Q

EMDB-30368: 
cryo_EM map of SLC26A9
Method: single particle / : Chi XM, Chen Y

EMDB-9834: 
Structure of RyR2 (F/A/C/L-Ca2+/apo-CaM-M dataset)
Method: single particle / : Gong DS, Chi XM

EMDB-9836: 
Structure of RyR2 (F/A/C/L-Ca2+/Ca2+CaM dataset)
Method: single particle / : Gong DS, Chi XM

EMDB-9837: 
Structure of RyR2 (F/A/C/H-Ca2+/Ca2+CaM dataset)
Method: single particle / : Gong DS, Chi XM

EMDB-9880: 
Structure of RyR2 (*F/A/C/L-Ca2+/Ca2+-CaM dataset)
Method: single particle / : Gong DS, Chi XM

EMDB-9718: 
The crosslinked complex of ISWI-nucleosome in the ADP.BeF-bound state
Method: single particle / : Yan LJ, Wu H, Li XM, Gao N, Chen ZC

EMDB-9719: 
The complex of ISWI-nucleosome in the ADP.BeF-bound state
Method: single particle / : Yan LJ, Wu H, Li XM, Gao N, Chen ZC

PDB-5j0n: 
Lambda excision HJ intermediate
Method: single particle / : Van Duyne G, Grigorieff N, Landy A

EMDB-3400: 
Lambda excision HJ intermediate
Method: single particle / : Laxmikanthan G, Xu C, Brilot AF, Warren D, Steele L, Seah N, Tong W, Grigorieff N, Landy A, Van Duyne G
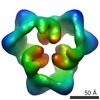
EMDB-1897: 
Reconstruction of the 3D model of AMPK trimer in basal state
Method: single particle / : Zhu L, Chen L, Zhou XM, Zhang YY, Zhang YJ, Zhao J, Ji SR, Wu JW, Wu Y
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search







 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
