[English] 日本語
 Yorodumi
Yorodumi- EMDB-63995: The structure of mCAT1 in complex with its substrate ornithine an... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of mCAT1 in complex with its substrate ornithine and the RBD of FrMLV. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | receptor / complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationL-ornithine transmembrane transporter activity / L-lysine transmembrane transporter activity / lysine transport / L-ornithine transmembrane transport / L-histidine import across plasma membrane / Amino acid transport across the plasma membrane / L-arginine transmembrane transport / L-histidine transmembrane transporter activity / L-arginine transmembrane transporter activity / positive regulation of T cell proliferation ...L-ornithine transmembrane transporter activity / L-lysine transmembrane transporter activity / lysine transport / L-ornithine transmembrane transport / L-histidine import across plasma membrane / Amino acid transport across the plasma membrane / L-arginine transmembrane transport / L-histidine transmembrane transporter activity / L-arginine transmembrane transporter activity / positive regulation of T cell proliferation / virus receptor activity / basolateral plasma membrane / apical plasma membrane / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / protein-containing complex / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |   Friend murine leukemia virus (ISOLATE 57) Friend murine leukemia virus (ISOLATE 57) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.65 Å | |||||||||
 Authors Authors | Xia LY / Yang Y / Chen XM | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Structural insights into cationic amino acid transport and viral receptor engagement by CAT1. Authors: Lingyun Xia / Bingqian Lin / Rongfeng Zou / Yanyan Wu / Jiaying Xu / Yan Pan / Yuan Yuan / Shuo Li / Yang Yang / Xuemin Chen /   Abstract: Cationic amino acid transporter 1 (CAT1) transports cationic amino acids and plays pivotal roles in cancer proliferation, immune modulation, and nitric oxide metabolism. It also serves as the ...Cationic amino acid transporter 1 (CAT1) transports cationic amino acids and plays pivotal roles in cancer proliferation, immune modulation, and nitric oxide metabolism. It also serves as the specific cellular receptor for certain murine leukemia viruses. Here, we report the cryo-electron microscopy (cryo-EM) structure of mammalian CAT1 in complex with its substrate ornithine and the receptor-binding domain (RBD) of Friend murine leukemia virus (FrMLV). CAT1 specifically recognizes the side-chain amino group of ornithine via residue S347 on transmembrane helix 8 (TM8), capturing the transporter in an inward-facing occluded conformation. Notably, the FrMLV RBD (frRBD) primarily engages the third extracellular loop (ECL3) of CAT1-a region marked by substantial species-specific variation that likely governs cross-species viral tropism. Together, our structural and biochemical results elucidate the molecular mechanism of substrate recognition and transport by mCAT1, and unveil the molecular basis for FrMLV receptor specificity. These findings provide a valuable framework for structure-based drug design targeting CAT1 in cancer and infectious diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_63995.map.gz emd_63995.map.gz | 57.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-63995-v30.xml emd-63995-v30.xml emd-63995.xml emd-63995.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_63995.png emd_63995.png | 37.4 KB | ||
| Filedesc metadata |  emd-63995.cif.gz emd-63995.cif.gz | 6.3 KB | ||
| Others |  emd_63995_half_map_1.map.gz emd_63995_half_map_1.map.gz emd_63995_half_map_2.map.gz emd_63995_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-63995 http://ftp.pdbj.org/pub/emdb/structures/EMD-63995 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63995 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63995 | HTTPS FTP |
-Related structure data
| Related structure data |  9uatMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_63995.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_63995.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_63995_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_63995_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The structure of mCAT1 in complex with its substrate ornithine an...
| Entire | Name: The structure of mCAT1 in complex with its substrate ornithine and the RBD of FrMLV |
|---|---|
| Components |
|
-Supramolecule #1: The structure of mCAT1 in complex with its substrate ornithine an...
| Supramolecule | Name: The structure of mCAT1 in complex with its substrate ornithine and the RBD of FrMLV type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: High affinity cationic amino acid transporter 1
| Macromolecule | Name: High affinity cationic amino acid transporter 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.978207 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGCKNLLGLG QQMLRRKVVD CSREESRLSR CLNTYDLVAL GVGSTLGAGV YVLAGAVARE NAGPAIVISF LIAALASVLA GLCYGEFGA RVPKTGSAYL YSYVTVGELW AFITGWNLIL SYIIGTSSVA RAWSATFDEL IGKPIGEFSR QHMALNAPGV L AQTPDIFA ...String: MGCKNLLGLG QQMLRRKVVD CSREESRLSR CLNTYDLVAL GVGSTLGAGV YVLAGAVARE NAGPAIVISF LIAALASVLA GLCYGEFGA RVPKTGSAYL YSYVTVGELW AFITGWNLIL SYIIGTSSVA RAWSATFDEL IGKPIGEFSR QHMALNAPGV L AQTPDIFA VIIIIILTGL LTLGVKESAM VNKIFTCINV LVLCFIVVSG FVKGSIKNWQ LTEKNFSCNN NDTNVKYGEG GF MPFGFSG VLSGAATCFY AFVGFDCIAT TGEEVKNPQK AIPVGIVASL LICFIAYFGV SAALTLMMPY FCLDIDSPLP GAF KHQGWE EAKYAVAIGS LCALSTSLLG SMFPMPRVIY AMAEDGLLFK FLAKINNRTK TPVIATVTSG AIAAVMAFLF ELKD LVDLM SIGTLLAYSL VAACVLVLRY QPEQPNLVYQ MARTTEELDR VDQNELVSAS ESQTGFLPVA EKFSLKSILS PKNVE PSKF SGLIVNISAG LLAALIITVC IVAVLGREAL AEGTLWAVFV MTGSVLLCML VTGIIWRQPE SKTKLSFKVP FVPVLP VLS IFVNIYLMMQ LDQGTWVRFA VWMLIGFTIY FGYGIWHS UniProtKB: High affinity cationic amino acid transporter 1 |
-Macromolecule #2: Surface protein
| Macromolecule | Name: Surface protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Friend murine leukemia virus (ISOLATE 57) Friend murine leukemia virus (ISOLATE 57) |
| Molecular weight | Theoretical: 25.411256 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: APGSSPHQVY NITWEVTNGD RETVWAISGN HPLWTWWPVL TPDLCMLALS GPPHWGLEYQ APYSSPPGPP CCSGSSGSSA GCSRDCDEP LTSLTPRCNT AWNRLKLDQV THKSSEGFYV CPGSHRPREA KSCGGPDSFY CASWGCETTG RVYWKPSSSW D YITVDNNL ...String: APGSSPHQVY NITWEVTNGD RETVWAISGN HPLWTWWPVL TPDLCMLALS GPPHWGLEYQ APYSSPPGPP CCSGSSGSSA GCSRDCDEP LTSLTPRCNT AWNRLKLDQV THKSSEGFYV CPGSHRPREA KSCGGPDSFY CASWGCETTG RVYWKPSSSW D YITVDNNL TTSQAVQVCK DNKWCNPLAI QFTNAGKQVT SWTTGHYWGL RLYVSGRDPG LTFGIRLRYQ N UniProtKB: Envelope glycoprotein |
-Macromolecule #5: L-ornithine
| Macromolecule | Name: L-ornithine / type: ligand / ID: 5 / Number of copies: 1 / Formula: ORN |
|---|---|
| Molecular weight | Theoretical: 132.161 Da |
| Chemical component information | 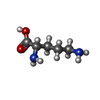 ChemComp-ORN: |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































