-Search query
-Search result
Showing 1 - 50 of 55 items for (author: bravo & jpk)
EMDB-41785: 
DdmD dimer in complex with ssDNA
EMDB-41790: 
DdmD monomer in complex with ssDNA

EMDB-41865: 
DdmDE handover complex
EMDB-41781: 
DdmE in complex with guide and target DNA

EMDB-44825: 
DdmD dimer apoprotein (CASP target)
EMDB-41088: 
SpRY-Cas9:gRNA complex bound to non-target DNA with 10 bp R-loop

EMDB-40681: 
SpRY-Cas9:gRNA complex targeting TGG PAM DNA

EMDB-40705: 
SpRY-Cas9:gRNA complex targeting TTC PAM DNA
EMDB-40740: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA

EMDB-41073: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 0 bp R-loop
EMDB-41074: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 2 bp R-loop

EMDB-41079: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 10 bp R-loop
EMDB-41080: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 13 bp R-loop
EMDB-41083: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 18 bp R-loop
EMDB-41084: 
SpRY-Cas9:gRNA complex bound to non-target DNA with 1 bp R-loop

EMDB-41085: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 3 bp R-loop
EMDB-41086: 
SpRY-Cas9:gRNA complex bound to non-target DNA with 6 bp R-loop
EMDB-41087: 
SpRY-Cas9:gRNA complex bound to non-target DNA with 8 bp R-loop
EMDB-41093: 
SpRYmer bound to NAC PAM DNA

EMDB-41775: 
SpG Cas9 with NGC PAM DNA target

EMDB-41867: 
SpG Cas9 with NGG PAM DNA target
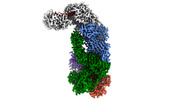
EMDB-40248: 
CRISPR-Cas type III-D effector complex
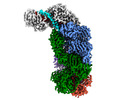
EMDB-40250: 
CRISPR-Cas type III-D effector complex bound to a self-target RNA in the pre-cleavage state
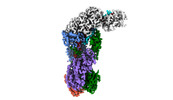
EMDB-40251: 
CRISPR-Cas type III-D effector complex bound to self-target RNA in a post-cleavage state

EMDB-40276: 
CRISPR-Cas type III-D effector complex consensus map

EMDB-40296: 
CRISPR-Cas type III-D effector complex local refinement map

EMDB-40297: 
CRISPR-Cas type III-D effector complex bound to a target RNA local refinement map

EMDB-40298: 
CRISPR-Cas type III-D effector complex bound to a target RNA consensus map
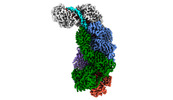
EMDB-40249: 
CRISPR-Cas type III-D effector complex bound to a target RNA

EMDB-41358: 
Structure of activated SAVED-CHAT filament

EMDB-27393: 
D. vulgaris type I-C Cascade bound to dsDNA target

EMDB-27403: 
type I-C Cascade bound to ssDNA target

EMDB-27412: 
type I-C Cascade bound to AcrIF2

EMDB-27402: 
type I-C Cascade

EMDB-27409: 
type I-C Cascade bound to AcrIC4

EMDB-27178: 
Structure of Cas12a2 binary complex
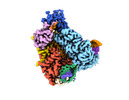
EMDB-27179: 
Structure of Cas12a2 ternary complex
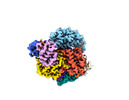
EMDB-27180: 
Structure of Cas12a2 ternary complex

EMDB-26485: 
Cryo-EM structure of pre-60S ribosomal subunit, unmethylated G2922
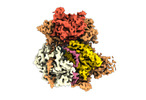
EMDB-24938: 
DrmAB:ADP
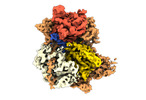
EMDB-24939: 
Structure of DrmAB:ADP:DNA complex
EMDB-24974: 
Structure of type I-D Cascade bound to a dsDNA target

EMDB-24976: 
Structure of type I-D Cascade bound to a ssRNA target

EMDB-24833: 
Cryo-EM structure of Cas9 in complex with 12-14MM DNA substrate, 5 minute time-point

EMDB-24834: 
Cas9 in complex with 12-14MM DNA at 60 minute time-point, linear conformation

EMDB-24835: 
Cas9 bound to 12-14MM DNA, 60 min time-point, kinked conformation

EMDB-24836: 
Cas9 in complex with 15-17MM DNA, 60 min time-point

EMDB-24837: 
Cas9 in complex with 18-20MM DNA, 1 min time-point, linear conformation

EMDB-24838: 
Cas9:gRNA in complex with 18-20MM DNA, 1 minute time-point, kinked active conformation

EMDB-13474: 
CryoEM structure of Rotavirus NSP2
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
