+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SpG Cas9 with NGG PAM DNA target | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SpG-Cas9 / CRISPR / Cas9 / IMMUNE SYSTEM / R-loop / IMMUNE SYSTEM-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / 3'-5' exonuclease activity / DNA endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / DNA binding / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Streptococcus pyogenes (bacteria) / synthetic construct (others) Streptococcus pyogenes (bacteria) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Bravo JPK / Hibshman GN / Taylor DW | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9. Authors: Grace N Hibshman / Jack P K Bravo / Matthew M Hooper / Tyler L Dangerfield / Hongshan Zhang / Ilya J Finkelstein / Kenneth A Johnson / David W Taylor /   Abstract: CRISPR-Cas9 is a powerful tool for genome editing, but the strict requirement for an NGG protospacer-adjacent motif (PAM) sequence immediately next to the DNA target limits the number of editable ...CRISPR-Cas9 is a powerful tool for genome editing, but the strict requirement for an NGG protospacer-adjacent motif (PAM) sequence immediately next to the DNA target limits the number of editable genes. Recently developed Cas9 variants have been engineered with relaxed PAM requirements, including SpG-Cas9 (SpG) and the nearly PAM-less SpRY-Cas9 (SpRY). However, the molecular mechanisms of how SpRY recognizes all potential PAM sequences remains unclear. Here, we combine structural and biochemical approaches to determine how SpRY interrogates DNA and recognizes target sites. Divergent PAM sequences can be accommodated through conformational flexibility within the PAM-interacting region, which facilitates tight binding to off-target DNA sequences. Nuclease activation occurs ~1000-fold slower than for Streptococcus pyogenes Cas9, enabling us to directly visualize multiple on-pathway intermediate states. Experiments with SpG position it as an intermediate enzyme between Cas9 and SpRY. Our findings shed light on the molecular mechanisms of PAMless genome editing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41867.map.gz emd_41867.map.gz | 483.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41867-v30.xml emd-41867-v30.xml emd-41867.xml emd-41867.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41867.png emd_41867.png | 51.7 KB | ||
| Filedesc metadata |  emd-41867.cif.gz emd-41867.cif.gz | 7.3 KB | ||
| Others |  emd_41867_half_map_1.map.gz emd_41867_half_map_1.map.gz emd_41867_half_map_2.map.gz emd_41867_half_map_2.map.gz | 474.6 MB 474.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41867 http://ftp.pdbj.org/pub/emdb/structures/EMD-41867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41867 | HTTPS FTP |
-Related structure data
| Related structure data |  8u3yMC  8spqC  8sqhC  8srsC  8t6oC  8t6pC  8t6sC  8t6tC  8t6xC  8t6yC  8t76C  8t77C  8t78C  8t79C  8t7sC  8tzzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41867.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41867.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.833 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_41867_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
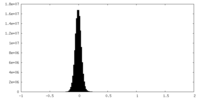
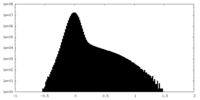
| Density Histograms |
-Half map: #2
| File | emd_41867_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
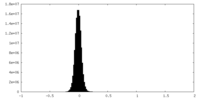
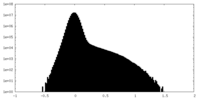
| Density Histograms |
- Sample components
Sample components
-Entire : SpG Cas9 with NGG PAM target
| Entire | Name: SpG Cas9 with NGG PAM target |
|---|---|
| Components |
|
-Supramolecule #1: SpG Cas9 with NGG PAM target
| Supramolecule | Name: SpG Cas9 with NGG PAM target / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 211.54 KDa |
-Macromolecule #1: CRISPR-associated endonuclease Cas9/Csn1
| Macromolecule | Name: CRISPR-associated endonuclease Cas9/Csn1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 158.476781 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KKYSIGLDIG TNSVGWAVIT DEYKVPSKKF KVLGNTDRHS IKKNLIGALL FDSGETAEAT RLKRTARRRY TRRKNRICYL QEIFSNEMA KVDDSFFHRL EESFLVEEDK KHERHPIFGN IVDEVAYHEK YPTIYHLRKK LVDSTDKADL RLIYLALAHM I KFRGHFLI ...String: KKYSIGLDIG TNSVGWAVIT DEYKVPSKKF KVLGNTDRHS IKKNLIGALL FDSGETAEAT RLKRTARRRY TRRKNRICYL QEIFSNEMA KVDDSFFHRL EESFLVEEDK KHERHPIFGN IVDEVAYHEK YPTIYHLRKK LVDSTDKADL RLIYLALAHM I KFRGHFLI EGDLNPDNSD VDKLFIQLVQ TYNQLFEENP INASGVDAKA ILSARLSKSR RLENLIAQLP GEKKNGLFGN LI ALSLGLT PNFKSNFDLA EDAKLQLSKD TYDDDLDNLL AQIGDQYADL FLAAKNLSDA ILLSDILRVN TEITKAPLSA SMI KRYDEH HQDLTLLKAL VRQQLPEKYK EIFFDQSKNG YAGYIDGGAS QEEFYKFIKP ILEKMDGTEE LLVKLNREDL LRKQ RTFDN GSIPHQIHLG ELHAILRRQE DFYPFLKDNR EKIEKILTFR IPYYVGPLAR GNSRFAWMTR KSEETITPWN FEEVV DKGA SAQSFIERMT NFDKNLPNEK VLPKHSLLYE YFTVYNELTK VKYVTEGMRK PAFLSGEQKK AIVDLLFKTN RKVTVK QLK EDYFKKIECF DSVEISGVED RFNASLGTYH DLLKIIKDKD FLDNEENEDI LEDIVLTLTL FEDREMIEER LKTYAHL FD DKVMKQLKRR RYTGWGRLSR KLINGIRDKQ SGKTILDFLK SDGFANRNFM QLIHDDSLTF KEDIQKAQVS GQGDSLHE H IANLAGSPAI KKGILQTVKV VDELVKVMGR HKPENIVIEM ARENQTTQKG QKNSRERMKR IEEGIKELGS QILKEHPVE NTQLQNEKLY LYYLQNGRDM YVDQELDINR LSDYDVDHIV PQSFLKDDSI DNKVLTRSDK NRGKSDNVPS EEVVKKMKNY WRQLLNAKL ITQRKFDNLT KAERGGLSEL DKAGFIKRQL VETRQITKHV AQILDSRMNT KYDENDKLIR EVKVITLKSK L VSDFRKDF QFYKVREINN YHHAHDAYLN AVVGTALIKK YPKLESEFVY GDYKVYDVRK MIAKSEQEIG KATAKYFFYS NI MNFFKTE ITLANGEIRK RPLIETNGET GEIVWDKGRD FATVRKVLSM PQVNIVKKTE VQTGGFSKES ILPKRNSDKL IAR KKDWDP KKYGGFLWPT VAYSVLVVAK VEKGKSKKLK SVKELLGITI MERSSFEKNP IDFLEAKGYK EVKKDLIIKL PKYS LFELE NGRKRMLASA KQLQKGNELA LPSKYVNFLY LASHYEKLKG SPEDNEQKQL FVEQHKHYLD EIIEQISEFS KRVIL ADAN LDKVLSAYNK HRDKPIREQA ENIIHLFTLT NLGAPAAFKY FDTTIDRKQY RSTKEVLDAT LIHQSITGLY ETRIDL SQL G UniProtKB: CRISPR-associated endonuclease Cas9/Csn1 |
-Macromolecule #2: RNA (98-MER)
| Macromolecule | Name: RNA (98-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 31.7029 KDa |
| Sequence | String: GACGCAUAAA GAUGAGACGC GUUUUAGAGC UAGAAAUAGC AAGUUAAAAU AAGGCUAGUC CGUUAUCAAC UUGAAAAAGU GGCACCGAG UCGGUGCUU |
-Macromolecule #3: DNA (5'-D(P*CP*GP*TP*TP*TP*GP*TP*AP*CP*TP*CP*CP*AP*GP*CP*G)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*GP*TP*TP*TP*GP*TP*AP*CP*TP*CP*CP*AP*GP*CP*G)-3') type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.865152 KDa |
| Sequence | String: (DC)(DG)(DT)(DT)(DT)(DG)(DT)(DA)(DC)(DT) (DC)(DC)(DA)(DG)(DC)(DG) |
-Macromolecule #4: DNA (5'-D(P*TP*CP*TP*CP*AP*TP*CP*TP*TP*TP*AP*TP*GP*CP*GP*TP*C)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*CP*TP*CP*AP*TP*CP*TP*TP*TP*AP*TP*GP*CP*GP*TP*C)-3') type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 5.119319 KDa |
| Sequence | String: (DT)(DC)(DT)(DC)(DA)(DT)(DC)(DT)(DT)(DT) (DA)(DT)(DG)(DC)(DG)(DT)(DC) |
-Macromolecule #5: DNA (5'-D(P*TP*GP*GP*AP*GP*TP*AP*CP*AP*AP*AP*CP*G)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*GP*GP*AP*GP*TP*AP*CP*AP*AP*AP*CP*G)-3') type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.024649 KDa |
| Sequence | String: (DT)(DG)(DG)(DA)(DG)(DT)(DA)(DC)(DA)(DA) (DA)(DC)(DG) |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 5 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)