[English] 日本語
 Yorodumi
Yorodumi- EMDB-8469: 3D cryo-EM reconstruction of nucleotide-free MsbA in lipid nanodisc -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8469 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3D cryo-EM reconstruction of nucleotide-free MsbA in lipid nanodisc | |||||||||
 Map data Map data | 3D cryo-EM reconstruction of nucleotide-free MsbA in lipid nanodisc | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type lipid A-core oligosaccharide transporter / ATPase-coupled lipid transmembrane transporter activity / ABC-type transporter activity /  ATP hydrolysis activity / ATP hydrolysis activity /  ATP binding / identical protein binding / ATP binding / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli (E. coli) / Escherichia coli (E. coli) /   Escherichia coli O157:H7 (bacteria) Escherichia coli O157:H7 (bacteria) | |||||||||
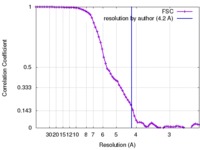
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.2 Å cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Mi W / Walz T / Liao M | |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Structural basis of MsbA-mediated lipopolysaccharide transport. Authors: Wei Mi / Yanyan Li / Sung Hwan Yoon / Robert K Ernst / Thomas Walz / Maofu Liao /   Abstract: Lipopolysaccharide (LPS) in the outer membrane of Gram-negative bacteria is critical for the assembly of their cell envelopes. LPS synthesized in the cytoplasmic leaflet of the inner membrane is ...Lipopolysaccharide (LPS) in the outer membrane of Gram-negative bacteria is critical for the assembly of their cell envelopes. LPS synthesized in the cytoplasmic leaflet of the inner membrane is flipped to the periplasmic leaflet by MsbA, an ATP-binding cassette transporter. Despite substantial efforts, the structural mechanisms underlying MsbA-driven LPS flipping remain elusive. Here we use single-particle cryo-electron microscopy to elucidate the structures of lipid-nanodisc-embedded MsbA in three functional states. The 4.2 Å-resolution structure of the transmembrane domains of nucleotide-free MsbA reveals that LPS binds deep inside MsbA at the height of the periplasmic leaflet, establishing extensive hydrophilic and hydrophobic interactions with MsbA. Two sub-nanometre-resolution structures of MsbA with ADP-vanadate and ADP reveal an unprecedented closed and an inward-facing conformation, respectively. Our study uncovers the structural basis for LPS recognition, delineates the conformational transitions of MsbA to flip LPS, and paves the way for structural characterization of other lipid flippases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8469.map.gz emd_8469.map.gz | 25.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8469-v30.xml emd-8469-v30.xml emd-8469.xml emd-8469.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8469_fsc.xml emd_8469_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_8469.png emd_8469.png | 56.2 KB | ||
| Others |  emd_8469_additional.map.gz emd_8469_additional.map.gz | 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8469 http://ftp.pdbj.org/pub/emdb/structures/EMD-8469 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8469 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8469 | HTTPS FTP |
-Related structure data
| Related structure data |  5tv4MC  8465C  8467C  8669C  8670C  8671C  5ttpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8469.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8469.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D cryo-EM reconstruction of nucleotide-free MsbA in lipid nanodisc | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: 3D cryo-EM reconstruction of nucleotide-free MsbA in lipid...
| File | emd_8469_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D cryo-EM reconstruction of nucleotide-free MsbA in lipid nanodisc, without low-pass filter or modification of amplitude | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MsbA reconstituted in lipid nanodisc
| Entire | Name: MsbA reconstituted in lipid nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: MsbA reconstituted in lipid nanodisc
| Supramolecule | Name: MsbA reconstituted in lipid nanodisc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)Recombinant plasmid: pET-19a |
-Macromolecule #1: Lipid A export ATP-binding/permease protein MsbA
| Macromolecule | Name: Lipid A export ATP-binding/permease protein MsbA / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number:  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances |
|---|---|
| Source (natural) | Organism:   Escherichia coli O157:H7 (bacteria) Escherichia coli O157:H7 (bacteria) |
| Molecular weight | Theoretical: 67.310445 KDa |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: MGHHHHHHHH HHSSGHIDDD DKHMHNDKDL STWQTFRRLW PTIAPFKAGL IVAGVALILN AASDTFMLSL LKPLLDDGFG KTDRSVLVW MPLVVIGLMI LRGITSYVSS YCISWVSGKV VMTMRRRLFG HMMGMPVSFF DKQSTGTLLS RITYDSEQVA S SSSGALIT ...String: MGHHHHHHHH HHSSGHIDDD DKHMHNDKDL STWQTFRRLW PTIAPFKAGL IVAGVALILN AASDTFMLSL LKPLLDDGFG KTDRSVLVW MPLVVIGLMI LRGITSYVSS YCISWVSGKV VMTMRRRLFG HMMGMPVSFF DKQSTGTLLS RITYDSEQVA S SSSGALIT VVREGASIIG LFIMMFYYSW QLSIILIVLA PIVSIAIRVV SKRFRNISKN MQNTMGQVTT SAEQMLKGHK EV LIFGGQE VETKRFDKVS NRMRLQGMKM VSASSISDPI IQLIASLALA FVLYAASFPS VMDSLTAGTI TVVFSSMIAL MRP LKSLTN VNAQFQRGMA ACQTLFTILD SEQEKDEGKR VIERATGDVE FRNVTFTYPG RDVPALRNIN LKIPAGKTVA LVGR SGSGK STIASLITRF YDIDEGEILM DGHDLREYTL ASLRNQVALV SQNVHLFNDT VANNIAYART EQYSREQIEE AARMA YAMD FINKMDNGLD TVIGENGVLL SGGQRQRIAI ARALLRDSPI LILDEATSAL DTESERAIQA ALDELQKNRT SLVIAH RLS TIEKADEIVV VEDGVIVERG THNDLLEHRG VYAQLHKMQF GQ |
-Macromolecule #3: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Macromolecule #4: 3-HYDROXY-TETRADECANOIC ACID
| Macromolecule | Name: 3-HYDROXY-TETRADECANOIC ACID / type: ligand / ID: 4 / Number of copies: 4 / Formula: FTT |
|---|---|
| Molecular weight | Theoretical: 244.37 Da |
| Chemical component information |  ChemComp-FTT: |
-Macromolecule #5: MYRISTIC ACID
| Macromolecule | Name: MYRISTIC ACID / type: ligand / ID: 5 / Number of copies: 1 / Formula: MYR |
|---|---|
| Molecular weight | Theoretical: 228.371 Da |
| Chemical component information |  ChemComp-MYR: |
-Macromolecule #6: LAURIC ACID
| Macromolecule | Name: LAURIC ACID / type: ligand / ID: 6 / Number of copies: 1 / Formula: DAO |
|---|---|
| Molecular weight | Theoretical: 200.318 Da |
| Chemical component information |  ChemComp-DAO: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 47.0 e/Å2 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X