[English] 日本語
 Yorodumi
Yorodumi- EMDB-9940: Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E2P... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9940 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E2P state class3) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
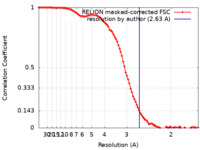
| Method | single particle reconstruction / cryo EM / Resolution: 2.63 Å | |||||||||
 Authors Authors | Hiraizumi M / Yamashita K / Nishizawa T / Nureki O | |||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Authors: Masahiro Hiraizumi / Keitaro Yamashita / Tomohiro Nishizawa / Osamu Nureki /  Abstract: In eukaryotic membranes, type IV P-type adenosine triphosphatases (P4-ATPases) mediate the translocation of phospholipids from the outer to the inner leaflet and maintain lipid asymmetry, which is ...In eukaryotic membranes, type IV P-type adenosine triphosphatases (P4-ATPases) mediate the translocation of phospholipids from the outer to the inner leaflet and maintain lipid asymmetry, which is critical for membrane trafficking and signaling pathways. Here, we report the cryo-electron microscopy structures of six distinct intermediates of the human ATP8A1-CDC50a heterocomplex at resolutions of 2.6 to 3.3 angstroms, elucidating the lipid translocation cycle of this P4-ATPase. ATP-dependent phosphorylation induces a large rotational movement of the actuator domain around the phosphorylation site in the phosphorylation domain, accompanied by lateral shifts of the first and second transmembrane helices, thereby allowing phosphatidylserine binding. The phospholipid head group passes through the hydrophilic cleft, while the acyl chain is exposed toward the lipid environment. These findings advance our understanding of the flippase mechanism and the disease-associated mutants of P4-ATPases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9940.map.gz emd_9940.map.gz | 11.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9940-v30.xml emd-9940-v30.xml emd-9940.xml emd-9940.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9940_fsc.xml emd_9940_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_9940.png emd_9940.png | 52.5 KB | ||
| Masks |  emd_9940_msk_1.map emd_9940_msk_1.map | 67 MB |  Mask map Mask map | |
| Others |  emd_9940_half_map_1.map.gz emd_9940_half_map_1.map.gz emd_9940_half_map_2.map.gz emd_9940_half_map_2.map.gz | 52.2 MB 52.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9940 http://ftp.pdbj.org/pub/emdb/structures/EMD-9940 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9940 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9940 | HTTPS FTP |
-Validation report
| Summary document |  emd_9940_validation.pdf.gz emd_9940_validation.pdf.gz | 489.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9940_full_validation.pdf.gz emd_9940_full_validation.pdf.gz | 489.4 KB | Display | |
| Data in XML |  emd_9940_validation.xml.gz emd_9940_validation.xml.gz | 15.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9940 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9940 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9940 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9940 | HTTPS FTP |
-Related structure data
| Related structure data |  9931C  9932C  9933C  9934C  9935C  9936C  9937C  9938C  9939C  9941C  9942C  6k7gC  6k7hC  6k7iC  6k7jC  6k7kC  6k7lC  6k7mC  6k7nC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10303 (Title: Cryo-EM structures of human P4-ATPase flippase / Data size: 11.0 TB EMPIAR-10303 (Title: Cryo-EM structures of human P4-ATPase flippase / Data size: 11.0 TBData #1: Unaligned movies for E2P class 1,2,3 [micrographs - multiframe] Data #2: Unaligned movies for E2Pi-PL and E1P [micrographs - multiframe] Data #3: Unaligned movies for E1P-ADP [micrographs - multiframe] Data #4: Unaligned movies for E1 class1,2,3 [micrographs - multiframe] Data #5: Unaligned movies for E1-ATP class 1,2,3 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9940.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9940.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9940_msk_1.map emd_9940_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_9940_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_9940_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ATP8A1-CDC50a
| Entire | Name: ATP8A1-CDC50a |
|---|---|
| Components |
|
-Supramolecule #1: ATP8A1-CDC50a
| Supramolecule | Name: ATP8A1-CDC50a / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant plasmid: pcDNA3.4 Homo sapiens (human) / Recombinant plasmid: pcDNA3.4 |
-Macromolecule #1: ATP8A1
| Macromolecule | Name: ATP8A1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GSREFMPTMR RTVSEIRSRA EGYEKTDDVS EKTSLADQEE VRTIFINQPQ LTKFCNNHVS TAKYNIITF LPRFLYSQFR RAANSFFLFI ALLQQIPDVS PTGRYTTLVP LLFILAVAAI K EIIEDIKR HKADNAVNKK QTQVLRNGAW EIVHWEKVNV GDIVIIKGKE ...String: GSREFMPTMR RTVSEIRSRA EGYEKTDDVS EKTSLADQEE VRTIFINQPQ LTKFCNNHVS TAKYNIITF LPRFLYSQFR RAANSFFLFI ALLQQIPDVS PTGRYTTLVP LLFILAVAAI K EIIEDIKR HKADNAVNKK QTQVLRNGAW EIVHWEKVNV GDIVIIKGKE YIPADTVLLS SS EPQAMCY IETSNLDGET NLKIRQGLPA TSDIKDVDSL MRISGRIECE SPNRHLYDFV GNI RLDGHG TVPLGADQIL LRGAQLRNTQ WVHGIVVYTG HDTKLMQNST SPPLKLSNVE RITN VQILI LFCILIAMSL VCSVGSAIWN RRHSGKDWYL NLNYGGASNF GLNFLTFIIL FNNLI PISL LVTLEVVKFT QAYFINWDLD MHYEPTDTAA MARTSNLNEE LGQVKYIFSD KTGTLT CNV MQFKKCTIAG VAYGQNSQFG DEKTFSDSSL LENLQNNHPT APIICEFLTM MAVCHTA VP ERERDKIIYQ AASPDEGALV RAAKQLNFVF TGRTPDSVII DSLGQEERYE LLNVLEFT S ARKRMSVIVR TPSGKLRLYC KGADTVIYDR LAETSKYKEI TLKHLEQFAT EGLRTLCFA VAEISESDFQ EWRAVYQRAS TSVQNRLLKL EESYELIEKN LQLLGATAIE DKLQDQVPET IETLMKADI KIWILTGDKQ ETAINIGHSC KLLKKNMGMI VINEGSLDGT RETLSRHCTT L GDALRKEN DFALIIDGKT LKYALTFGVR QYFLDLALSC KAVICCRVSP LQKSEVVEMV KK QVKVVTL AIGDGANDVS MIQTAHVGVG ISGNEGLQAA NSSDYSIAQF KYLKNLLMIH GAW NYNRVS KCILYCFYKN IVLYIIEIWF AFVNGFSGQI LFERWCIGLY NVMFTAMPPL TLGI FERSC RKENMLKYPE LYKTSQNALD FNTKVFWVHC LNGLFHSVIL FWFPLKALQY GTAFG NGKT SDYLLLGNFV YTFVVITVCL KAGLETSYWT WFSHIAIWGS IALWVVFFGI YSSLWP AIP MAPDMSGEAA MLFSSGVFWM GLLFIPVASL LLDVVYKVIK RTAFKTLVDE VQELEAK SQ DPGAVVLGKS LTERAQLLKN VFKKNHVNLY RSESLQQNLL HGYAFSQDEN GIVSQSEV I RAYDTTKQRP DEW |
-Macromolecule #2: CDC50a
| Macromolecule | Name: CDC50a / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAMNYNAKDE VDGGPPCAPG GTAKTRRPDN TAFKQQRLPA WQPILTAGTV LPIFFIIGLI FIPIGIGIF VTSNNIREIE IDYTGTEPSS PCNKCLSPDV TPCFCTINFT LEKSFEGNVF M YYGLSNFY QNHRRYVKSR DDSQLNGDSS ALLNPSKECE PYRRNEDKPI ...String: MAMNYNAKDE VDGGPPCAPG GTAKTRRPDN TAFKQQRLPA WQPILTAGTV LPIFFIIGLI FIPIGIGIF VTSNNIREIE IDYTGTEPSS PCNKCLSPDV TPCFCTINFT LEKSFEGNVF M YYGLSNFY QNHRRYVKSR DDSQLNGDSS ALLNPSKECE PYRRNEDKPI APCGAIANSM FN DTLELFL IGNDSYPIPI ALKKKGIAWW TDKNVKFRNP PGGDNLEERF KGTTKPVNWL KPV YMLDSD PDNNGFINED FIVWMRTAAL PTFRKLYRLI ERKSDLHPTL PAGRYSLNVT YNYP VHYFD GRKRMILSTI SWMGGKNPFL GIAYIAVGSI SFLLGVVLLV INHKYRNSSN TADIT I |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)