[English] 日本語
 Yorodumi
Yorodumi- EMDB-9937: Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9937 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ADP-Pi state) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | flippase / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationaminophospholipid translocation / positive regulation of phospholipid translocation / aminophospholipid flippase activity / chromaffin granule membrane / aminophospholipid transport / phosphatidylserine flippase activity / protein localization to endosome / phospholipid-translocating ATPase complex / phosphatidylserine floppase activity / positive regulation of protein exit from endoplasmic reticulum ...aminophospholipid translocation / positive regulation of phospholipid translocation / aminophospholipid flippase activity / chromaffin granule membrane / aminophospholipid transport / phosphatidylserine flippase activity / protein localization to endosome / phospholipid-translocating ATPase complex / phosphatidylserine floppase activity / positive regulation of protein exit from endoplasmic reticulum / ATPase-coupled intramembrane lipid transporter activity / ATPase-coupled monoatomic cation transmembrane transporter activity / xenobiotic transmembrane transport / P-type phospholipid transporter / phospholipid translocation / azurophil granule membrane / transport vesicle membrane / organelle membrane / Ion transport by P-type ATPases / synaptic vesicle endocytosis / specific granule membrane / transport across blood-brain barrier / learning / trans-Golgi network / positive regulation of neuron projection development / synaptic vesicle membrane / late endosome membrane / cytoplasmic vesicle / early endosome membrane / monoatomic ion transmembrane transport / apical plasma membrane / positive regulation of cell migration / intracellular membrane-bounded organelle / Neutrophil degranulation / glutamatergic synapse / structural molecule activity / magnesium ion binding / endoplasmic reticulum / Golgi apparatus / ATP hydrolysis activity / extracellular exosome / ATP binding / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
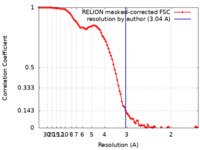
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Hiraizumi M / Yamashita K / Nishizawa T / Nureki O | |||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Authors: Masahiro Hiraizumi / Keitaro Yamashita / Tomohiro Nishizawa / Osamu Nureki /  Abstract: In eukaryotic membranes, type IV P-type adenosine triphosphatases (P4-ATPases) mediate the translocation of phospholipids from the outer to the inner leaflet and maintain lipid asymmetry, which is ...In eukaryotic membranes, type IV P-type adenosine triphosphatases (P4-ATPases) mediate the translocation of phospholipids from the outer to the inner leaflet and maintain lipid asymmetry, which is critical for membrane trafficking and signaling pathways. Here, we report the cryo-electron microscopy structures of six distinct intermediates of the human ATP8A1-CDC50a heterocomplex at resolutions of 2.6 to 3.3 angstroms, elucidating the lipid translocation cycle of this P4-ATPase. ATP-dependent phosphorylation induces a large rotational movement of the actuator domain around the phosphorylation site in the phosphorylation domain, accompanied by lateral shifts of the first and second transmembrane helices, thereby allowing phosphatidylserine binding. The phospholipid head group passes through the hydrophilic cleft, while the acyl chain is exposed toward the lipid environment. These findings advance our understanding of the flippase mechanism and the disease-associated mutants of P4-ATPases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9937.map.gz emd_9937.map.gz | 11.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9937-v30.xml emd-9937-v30.xml emd-9937.xml emd-9937.xml | 26.5 KB 26.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9937_fsc.xml emd_9937_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_9937.png emd_9937.png | 55.1 KB | ||
| Masks |  emd_9937_msk_1.map emd_9937_msk_1.map | 67 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-9937.cif.gz emd-9937.cif.gz | 8.1 KB | ||
| Others |  emd_9937_half_map_1.map.gz emd_9937_half_map_1.map.gz emd_9937_half_map_2.map.gz emd_9937_half_map_2.map.gz | 52.1 MB 52.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9937 http://ftp.pdbj.org/pub/emdb/structures/EMD-9937 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9937 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9937 | HTTPS FTP |
-Related structure data
| Related structure data |  6k7kMC  9931C  9932C  9933C  9934C  9935C  9936C  9938C  9939C  9940C  9941C  9942C  6k7gC  6k7hC  6k7iC  6k7jC  6k7lC  6k7mC  6k7nC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10303 (Title: Cryo-EM structures of human P4-ATPase flippase / Data size: 11.0 TB EMPIAR-10303 (Title: Cryo-EM structures of human P4-ATPase flippase / Data size: 11.0 TBData #1: Unaligned movies for E2P class 1,2,3 [micrographs - multiframe] Data #2: Unaligned movies for E2Pi-PL and E1P [micrographs - multiframe] Data #3: Unaligned movies for E1P-ADP [micrographs - multiframe] Data #4: Unaligned movies for E1 class1,2,3 [micrographs - multiframe] Data #5: Unaligned movies for E1-ATP class 1,2,3 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9937.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9937.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
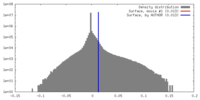
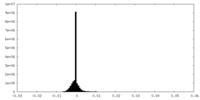
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9937_msk_1.map emd_9937_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
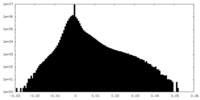
| Density Histograms |
-Half map: #2
| File | emd_9937_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_9937_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ATP8A1-CDC50a
| Entire | Name: ATP8A1-CDC50a |
|---|---|
| Components |
|
-Supramolecule #1: ATP8A1-CDC50a
| Supramolecule | Name: ATP8A1-CDC50a / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Phospholipid-transporting ATPase
| Macromolecule | Name: Phospholipid-transporting ATPase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: P-type phospholipid transporter |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 130.537852 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GSREFMPTMR RTVSEIRSRA EGYEKTDDVS EKTSLADQEE VRTIFINQPQ LTKFCNNHVS TAKYNIITFL PRFLYSQFRR AANSFFLFI ALLQQIPDVS PTGRYTTLVP LLFILAVAAI KEIIEDIKRH KADNAVNKKQ TQVLRNGAWE IVHWEKVNVG D IVIIKGKE ...String: GSREFMPTMR RTVSEIRSRA EGYEKTDDVS EKTSLADQEE VRTIFINQPQ LTKFCNNHVS TAKYNIITFL PRFLYSQFRR AANSFFLFI ALLQQIPDVS PTGRYTTLVP LLFILAVAAI KEIIEDIKRH KADNAVNKKQ TQVLRNGAWE IVHWEKVNVG D IVIIKGKE YIPADTVLLS SSEPQAMCYI ETSNLDGETN LKIRQGLPAT SDIKDVDSLM RISGRIECES PNRHLYDFVG NI RLDGHGT VPLGADQILL RGAQLRNTQW VHGIVVYTGH DTKLMQNSTS PPLKLSNVER ITNVQILILF CILIAMSLVC SVG SAIWNR RHSGKDWYLN LNYGGASNFG LNFLTFIILF NNLIPISLLV TLEVVKFTQA YFINWDLDMH YEPTDTAAMA RTSN LNEEL GQVKYIFSDK TGTLTCNVMQ FKKCTIAGVA YGQNSQFGDE KTFSDSSLLE NLQNNHPTAP IICEFLTMMA VCHTA VPER ERDKIIYQAA SPDEGALVRA AKQLNFVFTG RTPDSVIIDS LGQEERYELL NVLEFTSARK RMSVIVRTPS GKLRLY CKG ADTVIYDRLA ETSKYKEITL KHLEQFATEG LRTLCFAVAE ISESDFQEWR AVYQRASTSV QNRLLKLEES YELIEKN LQ LLGATAIEDK LQDQVPETIE TLMKADIKIW ILTGDKQETA INIGHSCKLL KKNMGMIVIN EGSLDGTRET LSRHCTTL G DALRKENDFA LIIDGKTLKY ALTFGVRQYF LDLALSCKAV ICCRVSPLQK SEVVEMVKKQ VKVVTLAIGD GANDVSMIQ TAHVGVGISG NEGLQAANSS DYSIAQFKYL KNLLMIHGAW NYNRVSKCIL YCFYKNIVLY IIEIWFAFVN GFSGQILFER WCIGLYNVM FTAMPPLTLG IFERSCRKEN MLKYPELYKT SQNALDFNTK VFWVHCLNGL FHSVILFWFP LKALQYGTAF G NGKTSDYL LLGNFVYTFV VITVCLKAGL ETSYWTWFSH IAIWGSIALW VVFFGIYSSL WPAIPMAPDM SGEAAMLFSS GV FWMGLLF IPVASLLLDV VYKVIKRTAF KTLVDEVQEL EAKSQDPGAV VLGKSLTERA QLLKNVFKKN HVNLYRSESL QQN LLHGYA FSQDENGIVS QSEVIRAYDT TKQRPDEW UniProtKB: Phospholipid-transporting ATPase |
-Macromolecule #2: Cell cycle control protein 50A
| Macromolecule | Name: Cell cycle control protein 50A / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.727527 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAMNYNAKDE VDGGPPCAPG GTAKTRRPDN TAFKQQRLPA WQPILTAGTV LPIFFIIGLI FIPIGIGIFV TSNNIREIEI DYTGTEPSS PCNKCLSPDV TPCFCTINFT LEKSFEGNVF MYYGLSNFYQ NHRRYVKSRD DSQLNGDSSA LLNPSKECEP Y RRNEDKPI ...String: MAMNYNAKDE VDGGPPCAPG GTAKTRRPDN TAFKQQRLPA WQPILTAGTV LPIFFIIGLI FIPIGIGIFV TSNNIREIEI DYTGTEPSS PCNKCLSPDV TPCFCTINFT LEKSFEGNVF MYYGLSNFYQ NHRRYVKSRD DSQLNGDSSA LLNPSKECEP Y RRNEDKPI APCGAIANSM FNDTLELFLI GNDSYPIPIA LKKKGIAWWT DKNVKFRNPP GGDNLEERFK GTTKPVNWLK PV YMLDSDP DNNGFINEDF IVWMRTAALP TFRKLYRLIE RKSDLHPTLP AGRYSLNVTY NYPVHYFDGR KRMILSTISW MGG KNPFLG IAYIAVGSIS FLLGVVLLVI NHKYRNSSNT ADITI UniProtKB: Cell cycle control protein 50A |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #5: TETRAFLUOROALUMINATE ION
| Macromolecule | Name: TETRAFLUOROALUMINATE ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: ALF |
|---|---|
| Molecular weight | Theoretical: 102.975 Da |
| Chemical component information |  ChemComp-ALF: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #8: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 8 / Number of copies: 1 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)