[English] 日本語
 Yorodumi
Yorodumi- EMDB-9750: Cryo-EM density map of E. coli 70S ribosome in complex with pepti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9750 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM density map of E. coli 70S ribosome in complex with peptide deformylase enzyme | |||||||||
 Map data Map data | Peptide deformylase (PDF) bound to E. coli 70S ribosome | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | E. coli 70S ribosome / pepdite deformylase / Nascent polypeptide exit tunnel / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationpeptide deformylase / peptide deformylase activity / : / ferrous iron binding / ribosome binding / hydrolase activity / translation / zinc ion binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
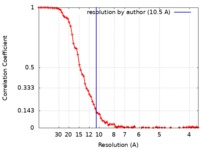
| Method | single particle reconstruction / cryo EM / Resolution: 10.5 Å | |||||||||
 Authors Authors | Sengupta J / Akbar S | |||||||||
| Funding support |  India, 2 items India, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2019 Journal: J Mol Biol / Year: 2019Title: Cryo-EM Structures Reveal Relocalization of MetAP in the Presence of Other Protein Biogenesis Factors at the Ribosomal Tunnel Exit. Authors: Sayan Bhakta / Shirin Akbar / Jayati Sengupta /  Abstract: During protein biosynthesis in bacteria, one of the earliest events that a nascent polypeptide chain goes through is the co-translational enzymatic processing. The event includes two enzymatic ...During protein biosynthesis in bacteria, one of the earliest events that a nascent polypeptide chain goes through is the co-translational enzymatic processing. The event includes two enzymatic pathways: deformylation of the N-terminal methionine by the enzyme peptide deformylase (PDF), followed by methionine excision catalyzed by methionine aminopeptidase (MetAP). During the enzymatic processing, the emerging nascent protein likely remains shielded by the ribosome-associated chaperone trigger factor. The ribosome tunnel exit serves as a stage for recruiting proteins involved in maturation processes of the nascent chain. Co-translational processing of nascent chains is a critical step for subsequent folding and functioning of mature proteins. Here, we present cryo-electron microscopy structures of Escherichia coli (E. coli) ribosome in complex with the nascent chain processing proteins. The structures reveal overlapping binding sites for PDF and MetAP when they bind individually at the tunnel exit site, where L22-L32 protein region provides primary anchoring sites for both proteins. In the absence of PDF, trigger factor can access ribosomal tunnel exit when MetAP occupies its primary binding site. Interestingly, however, in the presence of PDF, when MetAP's primary binding site is already engaged, MetAP has a remarkable ability to occupy an alternative binding site adjacent to PDF. Our study, thus, discloses an unexpected mechanism that MetAP adopts for context-specific ribosome association. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9750.map.gz emd_9750.map.gz | 37.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9750-v30.xml emd-9750-v30.xml emd-9750.xml emd-9750.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9750_fsc.xml emd_9750_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_9750.png emd_9750.png | 178.8 KB | ||
| Filedesc metadata |  emd-9750.cif.gz emd-9750.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9750 http://ftp.pdbj.org/pub/emdb/structures/EMD-9750 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9750 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9750 | HTTPS FTP |
-Validation report
| Summary document |  emd_9750_validation.pdf.gz emd_9750_validation.pdf.gz | 594.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9750_full_validation.pdf.gz emd_9750_full_validation.pdf.gz | 593.9 KB | Display | |
| Data in XML |  emd_9750_validation.xml.gz emd_9750_validation.xml.gz | 10.8 KB | Display | |
| Data in CIF |  emd_9750_validation.cif.gz emd_9750_validation.cif.gz | 13.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9750 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9750 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9750 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9750 | HTTPS FTP |
-Related structure data
| Related structure data |  6iy7MC  9752C  9753C  9759C  9778C  6iz7C  6iziC  6j0aC  6j45C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9750.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9750.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Peptide deformylase (PDF) bound to E. coli 70S ribosome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.89 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : E. coli 70S ribosome in complex with peptide deformylase
| Entire | Name: E. coli 70S ribosome in complex with peptide deformylase |
|---|---|
| Components |
|
-Supramolecule #1: E. coli 70S ribosome in complex with peptide deformylase
| Supramolecule | Name: E. coli 70S ribosome in complex with peptide deformylase type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Peptide deformylase
| Macromolecule | Name: Peptide deformylase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: peptide deformylase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.357447 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSVLQVLHIP DERLRKVAKP VEEVNAEIQR IVDDMFETMY AEEGIGLAAT QVDIHQRIIV IDVSENRDER LVLINPELLE KSGETGIEE GCLSIPEQRA LVPRAEKVKI RALDRDGKPF ELEADGLLAI CIQHEMDHLV GKLFMDYLSP LKQQRIRQKV E KLDRLKAR A UniProtKB: Peptide deformylase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 10.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)