+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9679 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tail structure of Vibrio phage M4 | |||||||||
 Map data Map data | Vibrio phage M4 contractile tail. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Viruses Viruses | |||||||||
| Method | helical reconstruction / negative staining / Resolution: 18.2 Å | |||||||||
 Authors Authors | Sen A / Das S / Dutta M / Ghosh AN | |||||||||
| Funding support |  India, 1 items India, 1 items
| |||||||||
 Citation Citation |  Journal: Arch Virol / Year: 2019 Journal: Arch Virol / Year: 2019Title: Structural analysis and proteomics studies on the Myoviridae vibriophage M4. Authors: Sayani Das / Moumita Dutta / Anindito Sen / Amar N Ghosh /   Abstract: Bacteriophages play a crucial role in tracking the spread of bacterial epidemics. The frequent emergence of antibiotic-resistant bacterial strains throughout the world has motivated studies on ...Bacteriophages play a crucial role in tracking the spread of bacterial epidemics. The frequent emergence of antibiotic-resistant bacterial strains throughout the world has motivated studies on bacteriophages that can potentially be used in phage therapy as an alternative to conventional antibiotic treatment. A recent outbreak of cholera in Haiti took many lives due to a rapid development of resistance to the available antibiotics. The properties of vibriophages, bacteriophages that infect Vibrio cholerae, are therefore of practical interest. A detailed understanding of the structure and assembly of a vibriophage is potentially useful in developing phage therapy against cholera as well as for fabricating artificial nanocontainers. Therefore, the aim of the present study was to determine the three-dimensional organization of vibriophage M4 at sub-nanometer resolution by electron microscopy and single-particle analysis techniques to facilitate its use as a therapeutic agent. We found that M4 has a large capsid with T = 13 icosahedral symmetry and a long contractile tail. This double-stranded DNA phage also contains a head-to-tail connector protein complex that joins the capsid to the tail and a prominent baseplate at the end of the tail. This study also provides information regarding the proteome of this phage, which is proteins similar to that of other Myoviridae phages, and most of the encoded proteins are structural proteins that form the exquisite architecture of this bacteriophage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9679.map.gz emd_9679.map.gz | 1.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9679-v30.xml emd-9679-v30.xml emd-9679.xml emd-9679.xml | 7.5 KB 7.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9679.png emd_9679.png | 115.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9679 http://ftp.pdbj.org/pub/emdb/structures/EMD-9679 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9679 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9679 | HTTPS FTP |
-Validation report
| Summary document |  emd_9679_validation.pdf.gz emd_9679_validation.pdf.gz | 78.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9679_full_validation.pdf.gz emd_9679_full_validation.pdf.gz | 77.5 KB | Display | |
| Data in XML |  emd_9679_validation.xml.gz emd_9679_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9679 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9679 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9679 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9679 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9679.map.gz / Format: CCP4 / Size: 6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9679.map.gz / Format: CCP4 / Size: 6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Vibrio phage M4 contractile tail. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
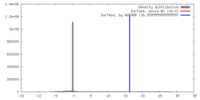
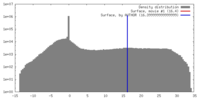
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Viruses
| Entire | Name:  Viruses Viruses |
|---|---|
| Components |
|
-Supramolecule #1: Viruses
| Supramolecule | Name: Viruses / type: virus / ID: 1 / Parent: 0 / Details: Vibrio phage M4, ATCC number-51354-B4 / NCBI-ID: 10239 / Sci species name: Viruses / Sci species strain: M4 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Vibrio cholerae MAK 757 (bacteria) Vibrio cholerae MAK 757 (bacteria) |
| Virus shell | Shell ID: 1 / Name: Vibrio phage M4 tail / Diameter: 240.0 Å |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Acetate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Average electron dose: 180.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 36.8 Å Applied symmetry - Helical parameters - Δ&Phi: 24.2 ° Applied symmetry - Helical parameters - Axial symmetry: C6 (6 fold cyclic) Resolution.type: BY AUTHOR / Resolution: 18.2 Å / Resolution method: FSC 0.33 CUT-OFF / Number images used: 800 |
|---|---|
| Final angle assignment | Type: COMMON LINE |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)