+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-7832 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | FLNaABD-Q170P bound to phalloidin-stabilized F-actin | |||||||||
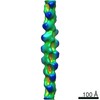
 マップデータ マップデータ | Symmetrized map of FLNaABD-Q170P bound to phalloidin-stabilized F-actin. This map has been low-pass filtered to 6.6 A and sharpened with a B-factor of -250. | |||||||||
 試料 試料 |
| |||||||||
| 生物種 |  Homo sapiens (ヒト) / Homo sapiens (ヒト) /  | |||||||||
| 手法 | らせん対称体再構成法 / クライオ電子顕微鏡法 / 解像度: 6.6 Å | |||||||||
 データ登録者 データ登録者 | Iwamoto DV / Huehn AR / Simon B / Huet-Calderwood C / Baldassarre M / Sindelar CV / Calderwood DA | |||||||||
 引用 引用 |  ジャーナル: Nat Struct Mol Biol / 年: 2018 ジャーナル: Nat Struct Mol Biol / 年: 2018タイトル: Structural basis of the filamin A actin-binding domain interaction with F-actin. 著者: Daniel V Iwamoto / Andrew Huehn / Bertrand Simon / Clotilde Huet-Calderwood / Massimiliano Baldassarre / Charles V Sindelar / David A Calderwood /   要旨: Actin-cross-linking proteins assemble actin filaments into higher-order structures essential for orchestrating cell shape, adhesion, and motility. Missense mutations in the tandem calponin homology ...Actin-cross-linking proteins assemble actin filaments into higher-order structures essential for orchestrating cell shape, adhesion, and motility. Missense mutations in the tandem calponin homology domains of their actin-binding domains (ABDs) underlie numerous genetic diseases, but a molecular understanding of these pathologies is hampered by the lack of high-resolution structures of any actin-cross-linking protein bound to F-actin. Here, taking advantage of a high-affinity, disease-associated mutant of the human filamin A (FLNa) ABD, we combine cryo-electron microscopy and functional studies to reveal at near-atomic resolution how the first calponin homology domain (CH1) and residues immediately N-terminal to it engage actin. We further show that reorientation of CH2 relative to CH1 is required to avoid clashes with actin and to expose F-actin-binding residues on CH1. Our data explain localization of disease-associated loss-of-function mutations to FLNaCH1 and gain-of-function mutations to the regulatory FLNaCH2. Sequence conservation argues that this provides a general model for ABD-F-actin binding. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_7832.map.gz emd_7832.map.gz | 20.8 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-7832-v30.xml emd-7832-v30.xml emd-7832.xml emd-7832.xml | 14.6 KB 14.6 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_7832_fsc.xml emd_7832_fsc.xml | 13.9 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_7832.png emd_7832.png | 34.1 KB | ||
| マスクデータ |  emd_7832_msk_1.map emd_7832_msk_1.map | 226.3 MB |  マスクマップ マスクマップ | |
| その他 |  emd_7832_half_map_1.map.gz emd_7832_half_map_1.map.gz emd_7832_half_map_2.map.gz emd_7832_half_map_2.map.gz | 32.6 MB 32.6 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7832 http://ftp.pdbj.org/pub/emdb/structures/EMD-7832 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7832 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7832 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_7832_validation.pdf.gz emd_7832_validation.pdf.gz | 79.3 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_7832_full_validation.pdf.gz emd_7832_full_validation.pdf.gz | 78.4 KB | 表示 | |
| XML形式データ |  emd_7832_validation.xml.gz emd_7832_validation.xml.gz | 495 B | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7832 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7832 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7832 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7832 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_7832.map.gz / 形式: CCP4 / 大きさ: 226.3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_7832.map.gz / 形式: CCP4 / 大きさ: 226.3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Symmetrized map of FLNaABD-Q170P bound to phalloidin-stabilized F-actin. This map has been low-pass filtered to 6.6 A and sharpened with a B-factor of -250. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.247 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-マスク #1
| ファイル |  emd_7832_msk_1.map emd_7832_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Independently refined half map(1) that was symmetrized with...
| ファイル | emd_7832_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Independently refined half map(1) that was symmetrized with parameters derived from the full map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Independently refined half map(2) that was symmetrized with...
| ファイル | emd_7832_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Independently refined half map(2) that was symmetrized with parameters derived from the full map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Helical complex of FLNaABD-Q170P bound to phalloidin-stabilized F...
| 全体 | 名称: Helical complex of FLNaABD-Q170P bound to phalloidin-stabilized F-actin |
|---|---|
| 要素 |
|
-超分子 #1: Helical complex of FLNaABD-Q170P bound to phalloidin-stabilized F...
| 超分子 | 名称: Helical complex of FLNaABD-Q170P bound to phalloidin-stabilized F-actin タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Filamin A Actin Binding Domain
| 分子 | 名称: Filamin A Actin Binding Domain / タイプ: protein_or_peptide / ID: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: PATEKDLAED APWKKIQQNT FTRWCNEHLK CVSKRIANLQ TDLSDGLRLI ALLEVLSQKK MHRKHNQRPT FRQMQLENVS VALEFLDRES IKLVSIDSKA IVDGNLKLIL GLIWTLILHY S |
-分子 #2: Actin
| 分子 | 名称: Actin / タイプ: protein_or_peptide / ID: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 配列 | 文字列: TTALVCDNGS GLVKAGFAGD DAPRAVFPSI VGRPRHQGVM VGMGQKDSYV GDEAQSKRGI LTLKYPIEHG IITNWDDMEK IWHHTFYNE LRVAPEEHPT LLTEAPLNPK ANREKMTQIM FETFNVPAMY VAIQAVLSLY ASGRTTGIVL DSGDGVTHNV P IYEGYALP ...文字列: TTALVCDNGS GLVKAGFAGD DAPRAVFPSI VGRPRHQGVM VGMGQKDSYV GDEAQSKRGI LTLKYPIEHG IITNWDDMEK IWHHTFYNE LRVAPEEHPT LLTEAPLNPK ANREKMTQIM FETFNVPAMY VAIQAVLSLY ASGRTTGIVL DSGDGVTHNV P IYEGYALP HAIMRLDLAG RDLTDYLMKI LTERGYSFVT TAEREIVRDI KEKLCYVALD FENEMATAAS SSSLEKSYEL PD GQVITIG NERFRCPETL FQPSFIGMES AGIHETTYNS IMKCDIDIRK DLYANNVMSG GTTMYPGIAD RMQKEITALA PST MKIKII APPERKYSVW IGGSILASLS TFQQMWITKQ EYDEAGPSIV HRKC |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | らせん対称体再構成法 |
| 試料の集合状態 | filament |
- 試料調製
試料調製
| 緩衝液 | pH: 8 |
|---|---|
| 凍結 | 凍結剤: ETHANE / 装置: HOMEMADE PLUNGER |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TECNAI F20 |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 平均電子線量: 60.0 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: SPOT SCAN / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Tecnai F20 / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)














































