+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7023 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mitochondrial ATPase Protease YME1 | ||||||||||||||||||||||||
 Map data Map data | sharpened map | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | Mitochondrial / ATPase / Protease / HYDROLASE | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationATP-dependent peptidase activity / metalloendopeptidase activity / ATP hydrolysis activity / mitochondrion / proteolysis / ATP binding / membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |    | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||||||||||||||
 Authors Authors | Puchades C / Rampello AJ | ||||||||||||||||||||||||
| Funding support |  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2017 Journal: Science / Year: 2017Title: Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Authors: Cristina Puchades / Anthony J Rampello / Mia Shin / Christopher J Giuliano / R Luke Wiseman / Steven E Glynn / Gabriel C Lander /  Abstract: We present an atomic model of a substrate-bound inner mitochondrial membrane AAA+ quality control protease in yeast, YME1. Our ~3.4-angstrom cryo-electron microscopy structure reveals how the ...We present an atomic model of a substrate-bound inner mitochondrial membrane AAA+ quality control protease in yeast, YME1. Our ~3.4-angstrom cryo-electron microscopy structure reveals how the adenosine triphosphatases (ATPases) form a closed spiral staircase encircling an unfolded substrate, directing it toward the flat, symmetric protease ring. Three coexisting nucleotide states allosterically induce distinct positioning of tyrosines in the central channel, resulting in substrate engagement and translocation to the negatively charged proteolytic chamber. This tight coordination by a network of conserved residues defines a sequential, around-the-ring adenosine triphosphate hydrolysis cycle that results in stepwise substrate translocation. A hingelike linker accommodates the large-scale nucleotide-driven motions of the ATPase spiral relative to the planar proteolytic base. The translocation mechanism is likely conserved for other AAA+ ATPases. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7023.map.gz emd_7023.map.gz | 11.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7023-v30.xml emd-7023-v30.xml emd-7023.xml emd-7023.xml | 33.6 KB 33.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7023_fsc.xml emd_7023_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_7023.png emd_7023.png | 199.9 KB | ||
| Filedesc metadata |  emd-7023.cif.gz emd-7023.cif.gz | 7.1 KB | ||
| Others |  emd_7023_additional_1.map.gz emd_7023_additional_1.map.gz emd_7023_additional_2.map.gz emd_7023_additional_2.map.gz emd_7023_additional_3.map.gz emd_7023_additional_3.map.gz emd_7023_half_map_1.map.gz emd_7023_half_map_1.map.gz emd_7023_half_map_2.map.gz emd_7023_half_map_2.map.gz | 1.7 MB 1.6 MB 4.4 MB 1.6 MB 1.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7023 http://ftp.pdbj.org/pub/emdb/structures/EMD-7023 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7023 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7023 | HTTPS FTP |
-Validation report
| Summary document |  emd_7023_validation.pdf.gz emd_7023_validation.pdf.gz | 780.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7023_full_validation.pdf.gz emd_7023_full_validation.pdf.gz | 780.4 KB | Display | |
| Data in XML |  emd_7023_validation.xml.gz emd_7023_validation.xml.gz | 13.5 KB | Display | |
| Data in CIF |  emd_7023_validation.cif.gz emd_7023_validation.cif.gz | 17.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7023 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7023 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7023 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7023 | HTTPS FTP |
-Related structure data
| Related structure data |  6az0MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7023.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7023.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
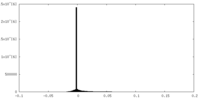
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: focused refinement of step subunit, unsharpened
| File | emd_7023_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused refinement of step subunit, unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
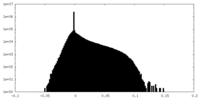
| Density Histograms |
-Additional map: focused refinement of step subunit, sharpened
| File | emd_7023_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused refinement of step subunit, sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
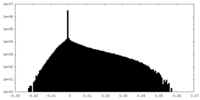
| Density Histograms |
-Additional map: unsharpened map
| File | emd_7023_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
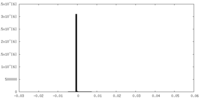
| Density Histograms |
-Half map: focused refinement of step subunit, half map 2
| File | emd_7023_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused refinement of step subunit, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: focused refinement of step subunit, half map 1
| File | emd_7023_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused refinement of step subunit, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : YME1 ATPase Protease Walker B mutant bound to substrate
| Entire | Name: YME1 ATPase Protease Walker B mutant bound to substrate |
|---|---|
| Components |
|
-Supramolecule #1: YME1 ATPase Protease Walker B mutant bound to substrate
| Supramolecule | Name: YME1 ATPase Protease Walker B mutant bound to substrate type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: YME1 Walker B mutant was recombinantly expressed in E. coli, solved in presence of ATP with unknown bound substrate. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 295 KDa |
-Macromolecule #1: Mitochondrial inner membrane i-AAA protease supercomplex subunit YME1
| Macromolecule | Name: Mitochondrial inner membrane i-AAA protease supercomplex subunit YME1 type: protein_or_peptide / ID: 1 / Details: Walker B mutant / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.025758 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KFDDVCGCDE ARAELEEIVD FLKDPTKYES LGGKLPKGVL LTGPPGTGKT LLARATAGEA GVDFFFMSGS EFDEVYVGVG AKRIRDLFA QARSRAPAII FIDQLDAIGG KRNPKDQAYA KQTLNQLLVE LDGFSQTSGI IIIGATNFPE ALDKALTRPG R FDKVVNVD ...String: KFDDVCGCDE ARAELEEIVD FLKDPTKYES LGGKLPKGVL LTGPPGTGKT LLARATAGEA GVDFFFMSGS EFDEVYVGVG AKRIRDLFA QARSRAPAII FIDQLDAIGG KRNPKDQAYA KQTLNQLLVE LDGFSQTSGI IIIGATNFPE ALDKALTRPG R FDKVVNVD LPDVRGRADI LKHHMKKITL ADNVDPTIIA RGTPGLSGAE LANLVNQAAV YACQKNAVSV DMSHFEWAKD KI LMGAERK TMVLTDAARK ATAFHEAGHA IMAKYTNGAT PLYKATILPR GRALGITFQL PEMDKVDITK RECQARLDVC MGG KIAEEL IYGKDNTTSG CGSDLQSATG TARAMVTQYG MSDDVGPVNL SEEWESWSNK IRDIADNEVI ELLKDSEERA RRLL TKKNV ELHRLAQGLI EYETLDAHEI EQVCKGEKLA KLKT UniProtKB: AAA+ ATPase domain-containing protein |
-Macromolecule #2: poly(UNK)
| Macromolecule | Name: poly(UNK) / type: protein_or_peptide / ID: 2 / Details: unknown polypeptide / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 869.063 Da |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 4 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 6 sec. / Pretreatment - Atmosphere: OTHER / Details: Gatan Solarus Plasma Cleaner | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Details | Data collected using Leginon |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-35 / Number grids imaged: 3 / Number real images: 6098 / Average exposure time: 7.0 sec. / Average electron dose: 60.0 e/Å2 Details: Images collected in counting mode at 5 frames per second. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.2 µm / Calibrated magnification: 51500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 100 |
|---|---|
| Output model |  PDB-6az0: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)