+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-6183 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | 3D structure of RepA-WH1 single filaments | |||||||||
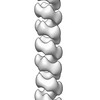
 マップデータ マップデータ | Negative-staining EM reconstruction of single repA-WH1 filament | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | RepA-WH1 prionoid / amyloid protofilaments / amyloid assembly | |||||||||
| 生物種 |  | |||||||||
| 手法 | らせん対称体再構成法 / ネガティブ染色法 / 解像度: 29.0 Å | |||||||||
 データ登録者 データ登録者 | Torreira E / Moreno M / Fuentes-Perez ME / Fernandez C / Martin-Benito J / Moreno-Herrero F / Giraldo R / Llorca O | |||||||||
 引用 引用 |  ジャーナル: Structure / 年: 2015 ジャーナル: Structure / 年: 2015タイトル: Amyloidogenesis of bacterial prionoid RepA-WH1 recapitulates dimer to monomer transitions of RepA in DNA replication initiation. 著者: Eva Torreira / María Moreno-Del Álamo / Maria Eugenia Fuentes-Perez / Cristina Fernández / Jaime Martín-Benito / Fernando Moreno-Herrero / Rafael Giraldo / Oscar Llorca /  要旨: Most available structures of amyloids correspond to peptide fragments that self-assemble in extended cross β sheets. However, structures in which a whole protein domain acts as building block of an ...Most available structures of amyloids correspond to peptide fragments that self-assemble in extended cross β sheets. However, structures in which a whole protein domain acts as building block of an amyloid fiber are scarce, in spite of their relevance to understand amyloidogenesis. Here, we use electron microscopy (EM) and atomic force microscopy (AFM) to analyze the structure of amyloid filaments assembled by RepA-WH1, a winged-helix domain from a DNA replication initiator in bacterial plasmids. RepA-WH1 functions as a cytotoxic bacterial prionoid that recapitulates features of mammalian amyloid proteinopathies. RepA are dimers that monomerize at the origin to initiate replication, and we find that RepA-WH1 reproduces this transition to form amyloids. RepA-WH1 assembles double helical filaments by lateral association of a single-stranded precursor built by monomers. Double filaments then associate in mature fibers. The intracellular and cytotoxic RepA-WH1 aggregates might reproduce the hierarchical assembly of human amyloidogenic proteins. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_6183.map.gz emd_6183.map.gz | 3 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-6183-v30.xml emd-6183-v30.xml emd-6183.xml emd-6183.xml | 10.3 KB 10.3 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_6183.jpg emd_6183.jpg | 456.9 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6183 http://ftp.pdbj.org/pub/emdb/structures/EMD-6183 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6183 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6183 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_6183_validation.pdf.gz emd_6183_validation.pdf.gz | 78.6 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_6183_full_validation.pdf.gz emd_6183_full_validation.pdf.gz | 77.7 KB | 表示 | |
| XML形式データ |  emd_6183_validation.xml.gz emd_6183_validation.xml.gz | 493 B | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6183 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6183 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6183 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6183 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_6183.map.gz / 形式: CCP4 / 大きさ: 3.3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_6183.map.gz / 形式: CCP4 / 大きさ: 3.3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Negative-staining EM reconstruction of single repA-WH1 filament | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 3.75 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Single RepA-WH1 filament
| 全体 | 名称: Single RepA-WH1 filament |
|---|---|
| 要素 |
|
-超分子 #1000: Single RepA-WH1 filament
| 超分子 | 名称: Single RepA-WH1 filament / タイプ: sample / ID: 1000 / 集合状態: single-chain helical filament / Number unique components: 1 |
|---|
-分子 #1: RepA
| 分子 | 名称: RepA / タイプ: protein_or_peptide / ID: 1 / 集合状態: single-chain helical filament / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  |
| 組換発現 | 生物種:  |
-実験情報
-構造解析
| 手法 | ネガティブ染色法 |
|---|---|
 解析 解析 | らせん対称体再構成法 |
| 試料の集合状態 | filament |
- 試料調製
試料調製
| 緩衝液 | pH: 8 詳細: 0.1 M Na2SO4, 40 mM HEPES, 5 mM MgSO4, 7% PEG4000, 3% MPD |
|---|---|
| 染色 | タイプ: NEGATIVE 詳細: Specimens were adsorbed on glow-discharged copper grids and stained using 2% uranyl acetate for 2 minutes. |
| グリッド | 詳細: 400-mesh copper grids |
| 凍結 | 凍結剤: NONE / 装置: OTHER |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | JEOL 1230 |
|---|---|
| アライメント法 | Legacy - 非点収差: Objective lens astigmatism was corrected using a TVIPS F416 CMOS and the EM-MENU software (TVIPS). |
| 日付 | 2012年9月1日 |
| 撮影 | カテゴリ: CCD フィルム・検出器のモデル: TVIPS TEMCAM-F416 (4k x 4k) デジタル化 - サンプリング間隔: 15.6 µm / 平均電子線量: 18 e/Å2 / ビット/ピクセル: 16 |
| 電子線 | 加速電圧: 100 kV / 電子線源: TUNGSTEN HAIRPIN |
| 電子光学系 | 倍率(補正後): 41586 / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 倍率(公称値): 41586 |
| 試料ステージ | 試料ホルダーモデル: JEOL |
- 画像解析
画像解析
| 詳細 | The particles were 2D-classified using CL2D implemented in Xmipp, and further processed using IHRSR. Note: Due to map resolution, the hand is arbitrary. |
|---|---|
| 最終 再構成 | 想定した対称性 - らせんパラメータ - Δz: 12.5 Å 想定した対称性 - らせんパラメータ - ΔΦ: 81 ° 想定した対称性 - らせんパラメータ - 軸対称性: C1 (非対称) 解像度のタイプ: BY AUTHOR / 解像度: 29.0 Å / 解像度の算出法: OTHER |
| CTF補正 | 詳細: Each micrograph using BSOFT |
-原子モデル構築 1
| 初期モデル | PDB ID: |
|---|---|
| ソフトウェア | 名称:  Chimera Chimera |
| 詳細 | Fitting was carried out both in individual segments and in the whole filament. In this case, helical symmetry was further applied to fill the EM map. |
| 精密化 | 空間: REAL / プロトコル: RIGID BODY FIT |
 ムービー
ムービー コントローラー
コントローラー









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















