+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5795 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
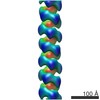
| Title | Helical reconstruction of hREGIIIalpha filaments on vesicles | |||||||||
 Map data Map data | Helical reconstruction of hREGIIIalpha filaments on vesicles | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | C-type lectin / membrane permeabilization / pore formation / hexameric pore / innate immunity / bactericidal toxin | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of detection of glucose / response to symbiotic bacterium / positive regulation of keratinocyte proliferation / negative regulation of keratinocyte differentiation / negative regulation of inflammatory response to wounding / oligosaccharide binding / peptidoglycan binding / heterophilic cell-cell adhesion / Antimicrobial peptides / positive regulation of wound healing ...positive regulation of detection of glucose / response to symbiotic bacterium / positive regulation of keratinocyte proliferation / negative regulation of keratinocyte differentiation / negative regulation of inflammatory response to wounding / oligosaccharide binding / peptidoglycan binding / heterophilic cell-cell adhesion / Antimicrobial peptides / positive regulation of wound healing / acute-phase response / hormone activity / response to wounding / response to peptide hormone / antimicrobial humoral immune response mediated by antimicrobial peptide / signaling receptor activity / carbohydrate binding / positive regulation of cell population proliferation / extracellular space / extracellular region / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 9.2 Å | |||||||||
 Authors Authors | Mukherjee S / Zheng H / Derebe M / Callenberg K / Partch CL / Rollins D / Propheter DC / Rizo J / Grabe M / Jiang Q-X / Hooper LV | |||||||||
 Citation Citation |  Journal: Nature / Year: 2014 Journal: Nature / Year: 2014Title: Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Authors: Sohini Mukherjee / Hui Zheng / Mehabaw G Derebe / Keith M Callenberg / Carrie L Partch / Darcy Rollins / Daniel C Propheter / Josep Rizo / Michael Grabe / Qiu-Xing Jiang / Lora V Hooper /  Abstract: Human body-surface epithelia coexist in close association with complex bacterial communities and are protected by a variety of antibacterial proteins. C-type lectins of the RegIII family are ...Human body-surface epithelia coexist in close association with complex bacterial communities and are protected by a variety of antibacterial proteins. C-type lectins of the RegIII family are bactericidal proteins that limit direct contact between bacteria and the intestinal epithelium and thus promote tolerance to the intestinal microbiota. RegIII lectins recognize their bacterial targets by binding peptidoglycan carbohydrate, but the mechanism by which they kill bacteria is unknown. Here we elucidate the mechanistic basis for RegIII bactericidal activity. We show that human RegIIIα (also known as HIP/PAP) binds membrane phospholipids and kills bacteria by forming a hexameric membrane-permeabilizing oligomeric pore. We derive a three-dimensional model of the RegIIIα pore by docking the RegIIIα crystal structure into a cryo-electron microscopic map of the pore complex, and show that the model accords with experimentally determined properties of the pore. Lipopolysaccharide inhibits RegIIIα pore-forming activity, explaining why RegIIIα is bactericidal for Gram-positive but not Gram-negative bacteria. Our findings identify C-type lectins as mediators of membrane attack in the mucosal immune system, and provide detailed insight into an antibacterial mechanism that promotes mutualism with the resident microbiota. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5795.map.gz emd_5795.map.gz | 1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5795-v30.xml emd-5795-v30.xml emd-5795.xml emd-5795.xml | 13.2 KB 13.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5795_1.jpg emd_5795_1.jpg | 94.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5795 http://ftp.pdbj.org/pub/emdb/structures/EMD-5795 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5795 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5795 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5795.map.gz / Format: CCP4 / Size: 6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5795.map.gz / Format: CCP4 / Size: 6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical reconstruction of hREGIIIalpha filaments on vesicles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.26 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : hREGIIIalpha filament
| Entire | Name: hREGIIIalpha filament |
|---|---|
| Components |
|
-Supramolecule #1000: hREGIIIalpha filament
| Supramolecule | Name: hREGIIIalpha filament / type: sample / ID: 1000 / Oligomeric state: Three-stranded helix / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 732 MDa / Theoretical: 880 MDa / Method: Calculated from sequence |
-Macromolecule #1: RegIIIalpha
| Macromolecule | Name: RegIIIalpha / type: protein_or_peptide / ID: 1 / Name.synonym: HIP/PAP Details: The purified protein was incubated with lipid vesicles to form the filaments. Number of copies: 1 / Oligomeric state: monomer in solution / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Tissue: Intestine / Cell: Enterocyte Homo sapiens (human) / synonym: Human / Tissue: Intestine / Cell: Enterocyte |
| Molecular weight | Experimental: 15 KDa / Theoretical: 15 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Regenerating islet-derived protein 3-alpha |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 5.5 / Details: 10 mM MES, 25 mM NaCl |
| Grid | Details: Quantifoil R2/2 200 mesh holey copper grids were covered with a layer of ultra-thin carbon film (1-3 nm) from the carbon side and were glow-discharged in a Denton Vacuum DV-502A instrument ...Details: Quantifoil R2/2 200 mesh holey copper grids were covered with a layer of ultra-thin carbon film (1-3 nm) from the carbon side and were glow-discharged in a Denton Vacuum DV-502A instrument with a 35 mA current for 60 s in amylamine atmosphere. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 92 % / Chamber temperature: 95.15 K / Instrument: FEI VITROBOT MARK III Method: Blot for 2 seconds before plunging into liquid ethane |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Temperature | Min: 93 K / Max: 103 K / Average: 100 K |
| Alignment procedure | Legacy - Astigmatism: corrected at 60,000x magnification |
| Specialist optics | Energy filter - Name: FEI / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 35.0 eV |
| Date | Dec 27, 2010 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 14 µm / Number real images: 75 / Average electron dose: 20 e/Å2 Details: The Zeiss SCAI scanner can handle six 4 x 5 inches Kodak SO-163 films at one time, so only films with no obvious drift or astigmatism were scanned. Od range: 1.5 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 61950 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder: Liquid nitrogen cooled / Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
| Details | IHRSR method was used. We could not determine handedness from tilt pairs as our microscope did not allow stable high-quality data collection. |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 18.52 Å Applied symmetry - Helical parameters - Δ&Phi: 54.21 ° Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.2 Å / Resolution method: OTHER / Software - Name: Spider Details: Final data were calculated from three separate datasets from three sessions of data collection. |
| CTF correction | Details: Each filament |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: Chimera, Situs |
| Details | The docking of one X-ray model into a segmented map corresponding to one subunit was first done manually in Chimera, and then optimized using Situs. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 20.76 / Target criteria: cross-correlation |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















