[English] 日本語
 Yorodumi
Yorodumi- EMDB-5920: Negative stain reconstruction of PGT151 Fab in complex with cleav... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5920 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain reconstruction of PGT151 Fab in complex with cleaved IAVI C22 Env trimer containing the transmembrane region | |||||||||
 Map data Map data | Negative stain reconstruction of IAVI C22 dCT Env trimer with PGT151 Fab bound | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 envelope glycoprotein trimer / broadly neutralizing antibody / PGT151 | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Lee JH / Blattner C / Wilson IA / Ward AB | |||||||||
 Citation Citation |  Journal: Immunity / Year: 2014 Journal: Immunity / Year: 2014Title: Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Authors: Claudia Blattner / Jeong Hyun Lee / Kwinten Sliepen / Ronald Derking / Emilia Falkowska / Alba Torrents de la Peña / Albert Cupo / Jean-Philippe Julien / Marit van Gils / Peter S Lee / ...Authors: Claudia Blattner / Jeong Hyun Lee / Kwinten Sliepen / Ronald Derking / Emilia Falkowska / Alba Torrents de la Peña / Albert Cupo / Jean-Philippe Julien / Marit van Gils / Peter S Lee / Wenjie Peng / James C Paulson / Pascal Poignard / Dennis R Burton / John P Moore / Rogier W Sanders / Ian A Wilson / Andrew B Ward /   Abstract: All previously characterized broadly neutralizing antibodies to the HIV-1 envelope glycoprotein (Env) target one of four major sites of vulnerability. Here, we define and structurally characterize a ...All previously characterized broadly neutralizing antibodies to the HIV-1 envelope glycoprotein (Env) target one of four major sites of vulnerability. Here, we define and structurally characterize a unique epitope on Env that is recognized by a recently discovered family of human monoclonal antibodies (PGT151-PGT158). The PGT151 epitope is comprised of residues and glycans at the interface of gp41 and gp120 within a single protomer and glycans from both subunits of a second protomer and represents a neutralizing epitope that is dependent on both gp120 and gp41. Because PGT151 binds only to properly formed, cleaved trimers, this distinctive property, and its ability to stabilize Env trimers, has enabled the successful purification of mature, cleaved Env trimers from the cell surface as a complex with PGT151. Here we compare the structural and functional properties of membrane-extracted Env trimers from several clades with those of the soluble, cleaved SOSIP gp140 trimer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
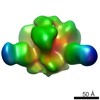
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5920.map.gz emd_5920.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5920-v30.xml emd-5920-v30.xml emd-5920.xml emd-5920.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| Images |  400_5920.gif 400_5920.gif 80_5920.gif 80_5920.gif | 28.4 KB 3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5920 http://ftp.pdbj.org/pub/emdb/structures/EMD-5920 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5920 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5920 | HTTPS FTP |
-Validation report
| Summary document |  emd_5920_validation.pdf.gz emd_5920_validation.pdf.gz | 77.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5920_full_validation.pdf.gz emd_5920_full_validation.pdf.gz | 77 KB | Display | |
| Data in XML |  emd_5920_validation.xml.gz emd_5920_validation.xml.gz | 492 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5920 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5920 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5920 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5920 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5920.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5920.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain reconstruction of IAVI C22 dCT Env trimer with PGT151 Fab bound | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
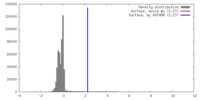
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Fab fragment of HIV-1 Env antibody PGT151 in complex with cleaved...
| Entire | Name: Fab fragment of HIV-1 Env antibody PGT151 in complex with cleaved IAVI C22 Env trimer with transmembrane region |
|---|---|
| Components |
|
-Supramolecule #1000: Fab fragment of HIV-1 Env antibody PGT151 in complex with cleaved...
| Supramolecule | Name: Fab fragment of HIV-1 Env antibody PGT151 in complex with cleaved IAVI C22 Env trimer with transmembrane region type: sample / ID: 1000 / Oligomeric state: 2 Fabs bind one Env Trimer / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 520 KDa |
-Macromolecule #1: HIV-1 Envelope glycoprotein
| Macromolecule | Name: HIV-1 Envelope glycoprotein / type: protein_or_peptide / ID: 1 / Name.synonym: Env Details: The trimer was co-expressed with Furin and Tat. Cleaved trimers were isolated using PGT151 antibody. Contains the transmembrane domain, but not the cytoplasmic domain. Number of copies: 1 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: IAVI C22 / synonym: HIV-1 Human immunodeficiency virus 1 / Strain: IAVI C22 / synonym: HIV-1 |
| Molecular weight | Theoretical: 420 MDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F / Recombinant plasmid: pSVIIIenv Homo sapiens (human) / Recombinant cell: HEK293F / Recombinant plasmid: pSVIIIenv |
-Macromolecule #2: PGT151 Antibody Fab fragment
| Macromolecule | Name: PGT151 Antibody Fab fragment / type: protein_or_peptide / ID: 2 / Name.synonym: PGT151 Fab / Details: Heavy chain-light chain dimer of PGT151 Fab / Number of copies: 2 / Oligomeric state: dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Cell: B-cells Homo sapiens (human) / synonym: Human / Cell: B-cells |
| Molecular weight | Theoretical: 49.5 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F Homo sapiens (human) / Recombinant cell: HEK293F |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50 mM Tris, 150 mM NaCl, 0.1% DDM |
| Staining | Type: NEGATIVE / Details: Sample diluted to <0.001% DDM, immediately applied to grid briefly, then stained for 30-45 seconds with 2% uranyl formate. |
| Grid | Details: plasma-cleaned carbon coated 400 mesh grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Temperature | Min: 293 K / Max: 293 K |
| Alignment procedure | Legacy - Astigmatism: Objective astigmatism corrected at 52,000 times magnification |
| Details | Data collected in 10 degree tilt increments |
| Date | Oct 19, 2013 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Sampling interval: 0.205 µm / Number real images: 1114 / Average electron dose: 26 e/Å2 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.55 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle max: 40 |
- Image processing
Image processing
| CTF correction | Details: not corrected |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: OTHER / Software - Name: XMIPP, EMAN, EMAN2, Sparx Details: Reference free 2D class averages generated using XMIPP and Sparx to sort particles. EMAN2 was used to generate an initial model followed by projection matching carried out in EMAN. Number images used: 6511 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: E / Chain - #3 - Chain ID: F / Chain - #4 - Chain ID: I / Chain - #5 - Chain ID: J |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | The PGV04 Fabs in the structure were removed prior to fitting. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - #0 - Chain ID: H / Chain - #1 - Chain ID: L |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Two Fabs were fit independently of each other into the EM reconstruction. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)