[English] 日本語
 Yorodumi
Yorodumi- EMDB-5874: Single-particle reconstruction of conformation XIV of ligand-free sGC -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5874 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single-particle reconstruction of conformation XIV of ligand-free sGC | |||||||||
 Map data Map data | Single-particle reconstruction of conformation XIV of ligand-free sGC | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | soluble guanylate cyclase / conformational heterogeneity | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 30.0 Å | |||||||||
 Authors Authors | Campbell MG / Underbakke ES / Potter CS / Carragher B / Marletta MA | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2014 Journal: Proc Natl Acad Sci U S A / Year: 2014Title: Single-particle EM reveals the higher-order domain architecture of soluble guanylate cyclase. Authors: Melody G Campbell / Eric S Underbakke / Clinton S Potter / Bridget Carragher / Michael A Marletta /  Abstract: Soluble guanylate cyclase (sGC) is the primary nitric oxide (NO) receptor in mammals and a central component of the NO-signaling pathway. The NO-signaling pathways mediate diverse physiological ...Soluble guanylate cyclase (sGC) is the primary nitric oxide (NO) receptor in mammals and a central component of the NO-signaling pathway. The NO-signaling pathways mediate diverse physiological processes, including vasodilation, neurotransmission, and myocardial functions. sGC is a heterodimer assembled from two homologous subunits, each comprised of four domains. Although crystal structures of isolated domains have been reported, no structure is available for full-length sGC. We used single-particle electron microscopy to obtain the structure of the complete sGC heterodimer and determine its higher-order domain architecture. Overall, the protein is formed of two rigid modules: the catalytic dimer and the clustered Per/Art/Sim and heme-NO/O2-binding domains, connected by a parallel coiled coil at two hinge points. The quaternary assembly demonstrates a very high degree of flexibility. We captured hundreds of individual conformational snapshots of free sGC, NO-bound sGC, and guanosine-5'-[(α,β)-methylene]triphosphate-bound sGC. The molecular architecture and pronounced flexibility observed provides a significant step forward in understanding the mechanism of NO signaling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5874.map.gz emd_5874.map.gz | 16.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5874-v30.xml emd-5874-v30.xml emd-5874.xml emd-5874.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5874.jpg emd_5874.jpg | 50.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5874 http://ftp.pdbj.org/pub/emdb/structures/EMD-5874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5874 | HTTPS FTP |
-Validation report
| Summary document |  emd_5874_validation.pdf.gz emd_5874_validation.pdf.gz | 77.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5874_full_validation.pdf.gz emd_5874_full_validation.pdf.gz | 77 KB | Display | |
| Data in XML |  emd_5874_validation.xml.gz emd_5874_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5874 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5874 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5874 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5874 | HTTPS FTP |
-Related structure data
| Related structure data | 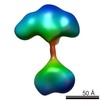 5861C 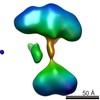 5862C  5863C 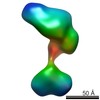 5864C  5865C  5866C  5867C  5868C 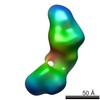 5869C 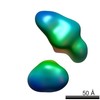 5870C 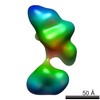 5871C 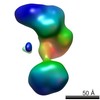 5872C 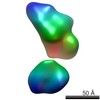 5873C  5875C 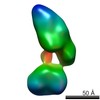 5876C  5877C 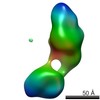 5878C 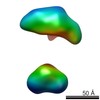 5879C 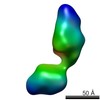 5880C 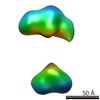 5881C  5882C 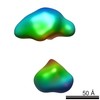 5883C  5884C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5874.map.gz / Format: CCP4 / Size: 41.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5874.map.gz / Format: CCP4 / Size: 41.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle reconstruction of conformation XIV of ligand-free sGC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Soluble Guanylate Cyclase, ligand-free
| Entire | Name: Soluble Guanylate Cyclase, ligand-free |
|---|---|
| Components |
|
-Supramolecule #1000: Soluble Guanylate Cyclase, ligand-free
| Supramolecule | Name: Soluble Guanylate Cyclase, ligand-free / type: sample / ID: 1000 / Oligomeric state: Heterodimer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 150 KDa / Theoretical: 150 KDa |
-Macromolecule #1: Soluble Guanylate Cyclase
| Macromolecule | Name: Soluble Guanylate Cyclase / type: protein_or_peptide / ID: 1 / Name.synonym: sGC / Number of copies: 1 / Oligomeric state: Heterodimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 150 KDa / Theoretical: 150 KDa |
| Recombinant expression | Organism:  Recombinant plasmid: pFastBac1/sGCALPHA1 and pFastBac1/sGCBETA1 |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50 mM TEA, 150 mM NaCl, 5 mM DTT |
|---|---|
| Staining | Type: NEGATIVE Details: 3 microliters of sample were applied to grid. The specimen was stained twice with 2% uranyl formate, then allowed to air-dry. |
| Grid | Details: Glow discharged C-flat grid with 2-micron-diameter holes overlaid by thin 1.5 nm continuous carbon |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 298 K |
| Date | Jan 26, 2013 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 2204 / Average electron dose: 35 e/Å2 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 80000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle min: -55 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | See publication |
|---|---|
| CTF correction | Details: Each micrograph |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 30.0 Å / Resolution method: OTHER / Software - Name: SPIDER / Number images used: 431 |
| Final two d classification | Number classes: 1 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















