[English] 日本語
 Yorodumi
Yorodumi- EMDB-48381: Azotobacter vinelandii Reduced MoFeP (C1 symmetry) obtained using... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Azotobacter vinelandii Reduced MoFeP (C1 symmetry) obtained using the SPT Labtech chameleon of 5 mM sodium dithionite under Al's oil | ||||||||||||
 Map data Map data | Sharpened EM map of MoFeP from Azotobacter vinellandii (C1 symmetry) in the presence of 5 mM sodium dithionite under Al's oil | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Nitrogenase / FeMoCo / nitrogen / P-cluster / METAL BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationmolybdenum-iron nitrogenase complex / nitrogenase / nitrogenase activity / nitrogen fixation / iron-sulfur cluster binding / ATP binding / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.33 Å | ||||||||||||
 Authors Authors | Cook BD / Narehood SM / McGuire KL / Li Y / Tezcan FA / Herzik MA | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Preparation of oxygen-sensitive proteins for high-resolution cryoEM structure determination using blot-free vitrification. Authors: Brian D Cook / Sarah M Narehood / Kelly L McGuire / Yizhou Li / F Akif Tezcan / Mark A Herzik /  Abstract: High-quality grid preparation for single-particle cryogenic electron microscopy (cryoEM) remains a bottleneck for routinely obtaining high-resolution structures. The issues that arise from ...High-quality grid preparation for single-particle cryogenic electron microscopy (cryoEM) remains a bottleneck for routinely obtaining high-resolution structures. The issues that arise from traditional grid preparation workflows are particularly exacerbated for oxygen-sensitive proteins, including metalloproteins, whereby oxygen-induced damage and alteration of oxidation states can result in protein inactivation, denaturation, and/or aggregation. Indeed, 99% of the current structures in the EMBD were prepared aerobically and limited successes for anaerobic cryoEM grid preparation exist. Current practices for anaerobic grid preparation involve a vitrification device located in an anoxic chamber, which presents significant challenges including temperature and humidity control, optimization of freezing conditions, costs for purchase and operation, as well as accessibility. Here, we present a streamlined approach that allows for the vitrification of oxygen-sensitive proteins in reduced states using an automated blot-free grid vitrification device - the SPT Labtech chameleon. This robust workflow allows for high-resolution structure determination of dynamic, oxygen-sensitive proteins, of varying complexity and molecular weight. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_48381.map.gz emd_48381.map.gz | 203.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-48381-v30.xml emd-48381-v30.xml emd-48381.xml emd-48381.xml | 26.9 KB 26.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_48381_fsc.xml emd_48381_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_48381.png emd_48381.png | 76 KB | ||
| Masks |  emd_48381_msk_1.map emd_48381_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-48381.cif.gz emd-48381.cif.gz | 7.4 KB | ||
| Others |  emd_48381_additional_1.map.gz emd_48381_additional_1.map.gz emd_48381_additional_2.map.gz emd_48381_additional_2.map.gz emd_48381_half_map_1.map.gz emd_48381_half_map_1.map.gz emd_48381_half_map_2.map.gz emd_48381_half_map_2.map.gz | 108.5 MB 6.8 MB 200.6 MB 200.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-48381 http://ftp.pdbj.org/pub/emdb/structures/EMD-48381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48381 | HTTPS FTP |
-Validation report
| Summary document |  emd_48381_validation.pdf.gz emd_48381_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_48381_full_validation.pdf.gz emd_48381_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_48381_validation.xml.gz emd_48381_validation.xml.gz | 21.5 KB | Display | |
| Data in CIF |  emd_48381_validation.cif.gz emd_48381_validation.cif.gz | 28.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-48381 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-48381 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-48381 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-48381 | HTTPS FTP |
-Related structure data
| Related structure data |  9mlyMC  9cqmC  9cqnC  9cqoC  9cqpC  9cqqC  9cqrC  9cqsC  9cqtC  9cquC  9cqvC  9cqwC  9cqxC  9cqyC  9cqzC  9cr0C  9mlzC  9mm0C  9mm1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_48381.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_48381.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened EM map of MoFeP from Azotobacter vinellandii (C1 symmetry) in the presence of 5 mM sodium dithionite under Al's oil | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.735 Å | ||||||||||||||||||||||||||||||||||||
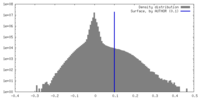
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_48381_msk_1.map emd_48381_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened EM map of MoFeP from Azotobacter vinellandii...
| File | emd_48381_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened EM map of MoFeP from Azotobacter vinellandii (C1 symmetry) in the presence of 5 mM sodium dithionite under Al's oil | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Resolve sharpened EM map of MoFeP from Azotobacter...
| File | emd_48381_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Resolve sharpened EM map of MoFeP from Azotobacter vinellandii (C1 symmetry) in the presence of 5 mM sodium dithionite under Al's oil | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B of MoFeP from Azotobacter vinellandii...
| File | emd_48381_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B of MoFeP from Azotobacter vinellandii (C1 symmetry) in the presence of 5 mM sodium dithionite under Al's oil | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B of MoFeP from Azotobacter vinellandii...
| File | emd_48381_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B of MoFeP from Azotobacter vinellandii (C1 symmetry) in the presence of 5 mM sodium dithionite under Al's oil | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Azotobacter vinelandii Reduced MoFeP (C1 symmetry) obtained using...
| Entire | Name: Azotobacter vinelandii Reduced MoFeP (C1 symmetry) obtained using the SPT Labtech chameleon of 5 mM sodium dithionite under Al's oil |
|---|---|
| Components |
|
-Supramolecule #1: Azotobacter vinelandii Reduced MoFeP (C1 symmetry) obtained using...
| Supramolecule | Name: Azotobacter vinelandii Reduced MoFeP (C1 symmetry) obtained using the SPT Labtech chameleon of 5 mM sodium dithionite under Al's oil type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) |
| Molecular weight | Theoretical: 230 KDa |
-Macromolecule #1: Nitrogenase molybdenum-iron protein alpha chain
| Macromolecule | Name: Nitrogenase molybdenum-iron protein alpha chain / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: nitrogenase |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) |
| Molecular weight | Theoretical: 55.363043 KDa |
| Sequence | String: MTGMSREEVE SLIQEVLEVY PEKARKDRNK HLAVNDPAVT QSKKCIISNK KSQPGLMTIR GCAYAGSKGV VWGPIKDMIH ISHGPVGCG QYSRAGRRNY YIGTTGVNAF VTMNFTSDFQ EKDIVFGGDK KLAKLIDEVE TLFPLNKGIS VQSECPIGLI G DDIESVSK ...String: MTGMSREEVE SLIQEVLEVY PEKARKDRNK HLAVNDPAVT QSKKCIISNK KSQPGLMTIR GCAYAGSKGV VWGPIKDMIH ISHGPVGCG QYSRAGRRNY YIGTTGVNAF VTMNFTSDFQ EKDIVFGGDK KLAKLIDEVE TLFPLNKGIS VQSECPIGLI G DDIESVSK VKGAELSKTI VPVRCEGFRG VSQSLGHHIA NDAVRDWVLG KRDEDTTFAS TPYDVAIIGD YNIGGDAWSS RI LLEEMGL RCVAQWSGDG SISEIELTPK VKLNLVHCYR SMNYISRHME EKYGIPWMEY NFFGPTKTIE SLRAIAAKFD ESI QKKCEE VIAKYKPEWE AVVAKYRPRL EGKRVMLYIG GLRPRHVIGA YEDLGMEVVG TGYEFAHNDD YDRTMKEMGD STLL YDDVT GYEFEEFVKR IKPDLIGSGI KEKFIFQKMG IPFREMHSWD YSGPYHGFDG FAIFARDMDM TLNNPCWKKL QAPWE ASEG AEKVAASA UniProtKB: Nitrogenase molybdenum-iron protein alpha chain |
-Macromolecule #2: Nitrogenase molybdenum-iron protein beta chain
| Macromolecule | Name: Nitrogenase molybdenum-iron protein beta chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: nitrogenase |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) |
| Molecular weight | Theoretical: 59.535879 KDa |
| Sequence | String: MSQQVDKIKA SYPLFLDQDY KDMLAKKRDG FEEKYPQDKI DEVFQWTTTK EYQELNFQRE ALTVNPAKAC QPLGAVLCAL GFEKTMPYV HGSQGCVAYF RSYFNRHFRE PVSCVSDSMT EDAAVFGGQQ NMKDGLQNCK ATYKPDMIAV STTCMAEVIG D DLNAFINN ...String: MSQQVDKIKA SYPLFLDQDY KDMLAKKRDG FEEKYPQDKI DEVFQWTTTK EYQELNFQRE ALTVNPAKAC QPLGAVLCAL GFEKTMPYV HGSQGCVAYF RSYFNRHFRE PVSCVSDSMT EDAAVFGGQQ NMKDGLQNCK ATYKPDMIAV STTCMAEVIG D DLNAFINN SKKEGFIPDE FPVPFAHTPS FVGSHVTGWD NMFEGIARYF TLKSMDDKVV GSNKKINIVP GFETYLGNFR VI KRMLSEM GVGYSLLSDP EEVLDTPADG QFRMYAGGTT QEEMKDAPNA LNTVLLQPWH LEKTKKFVEG TWKHEVPKLN IPM GLDWTD EFLMKVSEIS GQPIPASLTK ERGRLVDMMT DSHTWLHGKR FALWGDPDFV MGLVKFLLEL GCEPVHILCH NGNK RWKKA VDAILAASPY GKNATVYIGK DLWHLRSLVF TDKPDFMIGN SYGKFIQRDT LHKGKEFEVP LIRIGFPIFD RHHLH RSTT LGYEGAMQIL TTLVNSILER LDEETRGMQA TDYNHDLVR UniProtKB: Nitrogenase molybdenum-iron protein beta chain |
-Macromolecule #3: 3-HYDROXY-3-CARBOXY-ADIPIC ACID
| Macromolecule | Name: 3-HYDROXY-3-CARBOXY-ADIPIC ACID / type: ligand / ID: 3 / Number of copies: 2 / Formula: HCA |
|---|---|
| Molecular weight | Theoretical: 206.15 Da |
| Chemical component information |  ChemComp-HCA: |
-Macromolecule #4: iron-sulfur-molybdenum cluster with interstitial carbon
| Macromolecule | Name: iron-sulfur-molybdenum cluster with interstitial carbon type: ligand / ID: 4 / Number of copies: 2 / Formula: ICS |
|---|---|
| Molecular weight | Theoretical: 787.451 Da |
| Chemical component information |  ChemComp-ICE: |
-Macromolecule #5: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #6: FE(8)-S(7) CLUSTER
| Macromolecule | Name: FE(8)-S(7) CLUSTER / type: ligand / ID: 6 / Number of copies: 2 / Formula: CLF |
|---|---|
| Molecular weight | Theoretical: 671.215 Da |
| Chemical component information |  ChemComp-CLF: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 1037 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil Active R1.2/0.8 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 75 % / Chamber temperature: 298.15 K / Instrument: SPOTITON Details: Samples were frozen with the SPT Labtech chameleon. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Software | Name: EPU (ver. 2) |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 2501 / Average exposure time: 5.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 51 |
| Output model |  PDB-9mly: |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)