[English] 日本語
 Yorodumi
Yorodumi- PDB-9mm1: Azotobacter vinelandii Reduced MoFeP (C2 symmetry) obtained using... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9mm1 | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Azotobacter vinelandii Reduced MoFeP (C2 symmetry) obtained using the SPT Labtech chameleon of 60 mM sodium dithionite under Al's oil | |||||||||||||||||||||||||||||||||
 Components Components | (Nitrogenase molybdenum-iron protein ...) x 2 | |||||||||||||||||||||||||||||||||
 Keywords Keywords | METAL BINDING PROTEIN / Nitrogenase / FeMoCo / nitrogen / P-cluster | |||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmolybdenum-iron nitrogenase complex / nitrogenase / nitrogenase activity / nitrogen fixation / iron-sulfur cluster binding / ATP binding / metal ion binding Similarity search - Function | |||||||||||||||||||||||||||||||||
| Biological species |  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) | |||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.08 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Cook, B.D. / Narehood, S.M. / McGuire, K.L. / Li, Y. / Tezcan, F.A. / Herzik, M.A. | |||||||||||||||||||||||||||||||||
| Funding support |  United States, 3items United States, 3items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Preparation of oxygen-sensitive proteins for high-resolution cryoEM structure determination using blot-free vitrification. Authors: Brian D Cook / Sarah M Narehood / Kelly L McGuire / Yizhou Li / F Akif Tezcan / Mark A Herzik /  Abstract: High-quality grid preparation for single-particle cryogenic electron microscopy (cryoEM) remains a bottleneck for routinely obtaining high-resolution structures. The issues that arise from ...High-quality grid preparation for single-particle cryogenic electron microscopy (cryoEM) remains a bottleneck for routinely obtaining high-resolution structures. The issues that arise from traditional grid preparation workflows are particularly exacerbated for oxygen-sensitive proteins, including metalloproteins, whereby oxygen-induced damage and alteration of oxidation states can result in protein inactivation, denaturation, and/or aggregation. Indeed, 99% of the current structures in the EMBD were prepared aerobically and limited successes for anaerobic cryoEM grid preparation exist. Current practices for anaerobic grid preparation involve a vitrification device located in an anoxic chamber, which presents significant challenges including temperature and humidity control, optimization of freezing conditions, costs for purchase and operation, as well as accessibility. Here, we present a streamlined approach that allows for the vitrification of oxygen-sensitive proteins in reduced states using an automated blot-free grid vitrification device - the SPT Labtech chameleon. This robust workflow allows for high-resolution structure determination of dynamic, oxygen-sensitive proteins, of varying complexity and molecular weight. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9mm1.cif.gz 9mm1.cif.gz | 424.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9mm1.ent.gz pdb9mm1.ent.gz | 338.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9mm1.json.gz 9mm1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mm/9mm1 https://data.pdbj.org/pub/pdb/validation_reports/mm/9mm1 ftp://data.pdbj.org/pub/pdb/validation_reports/mm/9mm1 ftp://data.pdbj.org/pub/pdb/validation_reports/mm/9mm1 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  48384MC  9cqmC  9cqnC  9cqoC  9cqpC  9cqqC  9cqrC  9cqsC  9cqtC  9cquC  9cqvC  9cqwC  9cqxC  9cqyC  9cqzC  9cr0C  9mlyC  9mlzC  9mm0C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Nitrogenase molybdenum-iron protein ... , 2 types, 4 molecules ACBD
| #1: Protein | Mass: 55363.043 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Azotobacter vinelandii (bacteria) / References: UniProt: P07328, nitrogenase Azotobacter vinelandii (bacteria) / References: UniProt: P07328, nitrogenase#2: Protein | Mass: 59535.879 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Azotobacter vinelandii (bacteria) / References: UniProt: P07329, nitrogenase Azotobacter vinelandii (bacteria) / References: UniProt: P07329, nitrogenase |
|---|
-Non-polymers , 5 types, 1021 molecules 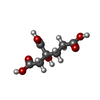
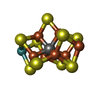



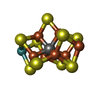



| #3: Chemical | | #4: Chemical | #5: Chemical | #6: Chemical | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Azotobacter vinelandii Reduced MoFeP (C2 symmetry) obtained using the SPT Labtech chameleon of 60 mM sodium dithionite under Al's oil Type: COMPLEX / Entity ID: #1-#2 / Source: NATURAL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.230 MDa / Experimental value: NO | |||||||||||||||
| Source (natural) | Organism:  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) | |||||||||||||||
| Buffer solution | pH: 7.5 | |||||||||||||||
| Buffer component |
| |||||||||||||||
| Specimen | Conc.: 8 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | |||||||||||||||
| Specimen support | Grid type: Quantifoil Active R1.2/0.8 | |||||||||||||||
| Vitrification | Instrument: SPT LABTECH CHAMELEON / Cryogen name: ETHANE / Humidity: 75 % / Chamber temperature: 298.15 K / Details: Samples were frozen with the SPT Labtech chameleon |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 165000 X / Nominal defocus max: 2500 nm / Nominal defocus min: 1000 nm / Cs: 2.7 mm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 5 sec. / Electron dose: 60 e/Å2 / Film or detector model: FEI FALCON IV (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 2000 |
| EM imaging optics | Energyfilter name: TFS Selectris X / Energyfilter slit width: 10 eV |
| Image scans | Width: 4096 / Height: 4096 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 678577 | ||||||||||||||||||||||||
| Symmetry | Point symmetry: C2 (2 fold cyclic) | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.08 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 105690 / Algorithm: FOURIER SPACE / Symmetry type: POINT | ||||||||||||||||||||||||
| Atomic model building | B value: 50.2 / Protocol: RIGID BODY FIT / Space: REAL | ||||||||||||||||||||||||
| Atomic model building | PDB-ID: 6NBC Accession code: 6NBC / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||
| Refinement | Highest resolution: 2.08 Å Stereochemistry target values: REAL-SPACE (WEIGHTED MAP SUM AT ATOM CENTERS) | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller




















 PDBj
PDBj





