[English] 日本語
 Yorodumi
Yorodumi- EMDB-45818: Human metHb (C2 symmetry) obtained using the SPT Labtech chameleon -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human metHb (C2 symmetry) obtained using the SPT Labtech chameleon | ||||||||||||
 Map data Map data | Sharpened EM map of metHb without Al's oil; C2 symmetry | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Hemoglobin / anaerobic / oxygen / heme / OXYGEN BINDING | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationHeme assimilation / nitric oxide transport / hemoglobin alpha binding / cellular oxidant detoxification / hemoglobin binding / haptoglobin-hemoglobin complex / renal absorption / hemoglobin complex / oxygen transport / Scavenging of heme from plasma ...Heme assimilation / nitric oxide transport / hemoglobin alpha binding / cellular oxidant detoxification / hemoglobin binding / haptoglobin-hemoglobin complex / renal absorption / hemoglobin complex / oxygen transport / Scavenging of heme from plasma / erythrocyte development / endocytic vesicle lumen / blood vessel diameter maintenance / oxygen carrier activity / hydrogen peroxide catabolic process / carbon dioxide transport / response to hydrogen peroxide / Heme signaling / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / Cytoprotection by HMOX1 / Late endosomal microautophagy / oxygen binding / platelet aggregation / regulation of blood pressure / Chaperone Mediated Autophagy / positive regulation of nitric oxide biosynthetic process / tertiary granule lumen / Factors involved in megakaryocyte development and platelet production / blood microparticle / ficolin-1-rich granule lumen / iron ion binding / inflammatory response / heme binding / Neutrophil degranulation / extracellular space / extracellular exosome / extracellular region / metal ion binding / membrane / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
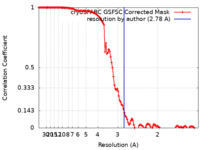
| Method | single particle reconstruction / cryo EM / Resolution: 2.78 Å | ||||||||||||
 Authors Authors | Cook BD / Narehood SM / McGuire KL / Li Y / Tezcan FA / Herzik MA | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Preparation of oxygen-sensitive proteins for high-resolution cryoEM structure determination using blot-free vitrification. Authors: Brian D Cook / Sarah M Narehood / Kelly L McGuire / Yizhou Li / F Akif Tezcan / Mark A Herzik /  Abstract: High-quality grid preparation for single-particle cryogenic electron microscopy (cryoEM) remains a bottleneck for routinely obtaining high-resolution structures. The issues that arise from ...High-quality grid preparation for single-particle cryogenic electron microscopy (cryoEM) remains a bottleneck for routinely obtaining high-resolution structures. The issues that arise from traditional grid preparation workflows are particularly exacerbated for oxygen-sensitive proteins, including metalloproteins, whereby oxygen-induced damage and alteration of oxidation states can result in protein inactivation, denaturation, and/or aggregation. Indeed, 99% of the current structures in the EMBD were prepared aerobically and limited successes for anaerobic cryoEM grid preparation exist. Current practices for anaerobic grid preparation involve a vitrification device located in an anoxic chamber, which presents significant challenges including temperature and humidity control, optimization of freezing conditions, costs for purchase and operation, as well as accessibility. Here, we present a streamlined approach that allows for the vitrification of oxygen-sensitive proteins in reduced states using an automated blot-free grid vitrification device - the SPT Labtech chameleon. This robust workflow allows for high-resolution structure determination of dynamic, oxygen-sensitive proteins, of varying complexity and molecular weight. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45818.map.gz emd_45818.map.gz | 156.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45818-v30.xml emd-45818-v30.xml emd-45818.xml emd-45818.xml | 27.8 KB 27.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45818_fsc.xml emd_45818_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_45818.png emd_45818.png | 43.3 KB | ||
| Masks |  emd_45818_msk_1.map emd_45818_msk_1.map | 166.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-45818.cif.gz emd-45818.cif.gz | 6.9 KB | ||
| Others |  emd_45818_additional_1.map.gz emd_45818_additional_1.map.gz emd_45818_additional_2.map.gz emd_45818_additional_2.map.gz emd_45818_half_map_1.map.gz emd_45818_half_map_1.map.gz emd_45818_half_map_2.map.gz emd_45818_half_map_2.map.gz | 83.1 MB 2.3 MB 154.4 MB 154.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45818 http://ftp.pdbj.org/pub/emdb/structures/EMD-45818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45818 | HTTPS FTP |
-Related structure data
| Related structure data |  9cqpMC  9cqmC  9cqnC  9cqoC  9cqqC  9cqrC  9cqsC  9cqtC  9cquC  9cqvC  9cqwC  9cqxC  9cqyC  9cqzC  9cr0C  9mlyC  9mlzC  9mm0C  9mm1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45818.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45818.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened EM map of metHb without Al's oil; C2 symmetry | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.735 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_45818_msk_1.map emd_45818_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened EM map of metHb without Al's oil; C2 symmetry
| File | emd_45818_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened EM map of metHb without Al's oil; C2 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: phenix RESOLVE modified EM map of metHb without Al's oil; C2 symmetry
| File | emd_45818_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | phenix RESOLVE modified EM map of metHb without Al's oil; C2 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B of metHb without Al's oil; C2 symmetry
| File | emd_45818_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of metHb without Al's oil; C2 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A of metHb without Al's oil; C2 symmetry
| File | emd_45818_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of metHb without Al's oil; C2 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human metHb (C2 symmetry) determined using the SPT Labtech chamel...
| Entire | Name: Human metHb (C2 symmetry) determined using the SPT Labtech chameleon without Al's oil |
|---|---|
| Components |
|
-Supramolecule #1: Human metHb (C2 symmetry) determined using the SPT Labtech chamel...
| Supramolecule | Name: Human metHb (C2 symmetry) determined using the SPT Labtech chameleon without Al's oil type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 64 KDa |
-Macromolecule #1: Hemoglobin subunit alpha
| Macromolecule | Name: Hemoglobin subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.993159 KDa |
| Sequence | String: VLSPADKTNV KAAWGKVGAH AGEYGAEALE RMFLSFPTTK TYFPHFDLSH GSAQVKGHGK KVADALTNAV AHVDDMPNAL SALSDLHAH KLRVDPVNFK LLSHCLLVTL AAHLPAEFTP AVHASLDKFL ASVSTVLTSK Y UniProtKB: Hemoglobin subunit alpha |
-Macromolecule #2: Hemoglobin subunit beta
| Macromolecule | Name: Hemoglobin subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.890198 KDa |
| Sequence | String: VHLTPEEKSA VTALWGKVNV DEVGGEALGR LLVVYPWTQR FFESFGDLST PDAVMGNPKV KAHGKKVLGA FSDGLAHLDN LKGTFATLS ELHCDKLHVD PENFRLLGNV LVCVLAHHFG KEFTPPVQAA YQKVVAGVAN ALAHKYH UniProtKB: Hemoglobin subunit beta |
-Macromolecule #3: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 3 / Number of copies: 4 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 75 % / Chamber temperature: 298.15 K / Instrument: SPT LABTECH CHAMELEON Details: Samples were frozen with the SPT Labtech chameleon. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Software | Name: EPU (ver. 2) |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 2847 / Average exposure time: 5.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)