[English] 日本語
 Yorodumi
Yorodumi- EMDB-4631: A dimer component of alpha-1 antitrypsin polymers isolated from Z... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4631 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | A dimer component of alpha-1 antitrypsin polymers isolated from ZZ explant liver tissue and decorated with Fab 4B12 (component B with ~90 degree rotation around the dimer axis) | ||||||||||||
 Map data Map data | Unmasked dimer component of alpha-1 antitrypsin polymers isolated from explant liver tissue; subunits rotated ~90 degrees around dimer axis. Partially resolved third subunit evident. | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||
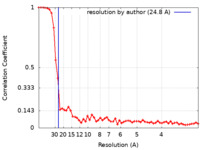
| Method | single particle reconstruction / negative staining / Resolution: 24.8 Å | ||||||||||||
 Authors Authors | Faull SV / Elliston ELK / Orlova EV / Gooptu B / Lomas DA / Irving JA | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2020 Journal: Sci Adv / Year: 2020Title: The structural basis for Z α-antitrypsin polymerization in the liver. Authors: Sarah V Faull / Emma L K Elliston / Bibek Gooptu / Alistair M Jagger / Ibrahim Aldobiyan / Adam Redzej / Magd Badaoui / Nina Heyer-Chauhan / S Tamir Rashid / Gary M Reynolds / David H Adams ...Authors: Sarah V Faull / Emma L K Elliston / Bibek Gooptu / Alistair M Jagger / Ibrahim Aldobiyan / Adam Redzej / Magd Badaoui / Nina Heyer-Chauhan / S Tamir Rashid / Gary M Reynolds / David H Adams / Elena Miranda / Elena V Orlova / James A Irving / David A Lomas /   Abstract: The serpinopathies are among a diverse set of conformational diseases that involve the aberrant self-association of proteins into ordered aggregates. α-Antitrypsin deficiency is the archetypal ...The serpinopathies are among a diverse set of conformational diseases that involve the aberrant self-association of proteins into ordered aggregates. α-Antitrypsin deficiency is the archetypal serpinopathy and results from the formation and deposition of mutant forms of α-antitrypsin as "polymer" chains in liver tissue. No detailed structural analysis has been performed of this material. Moreover, there is little information on the relevance of well-studied artificially induced polymers to these disease-associated molecules. We have isolated polymers from the liver tissue of Z α-antitrypsin homozygotes (E342K) who have undergone transplantation, labeled them using a Fab fragment, and performed single-particle analysis of negative-stain electron micrographs. The data show structural equivalence between heat-induced and ex vivo polymers and that the intersubunit linkage is best explained by a carboxyl-terminal domain swap between molecules of α-antitrypsin. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4631.map.gz emd_4631.map.gz | 12 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4631-v30.xml emd-4631-v30.xml emd-4631.xml emd-4631.xml | 22.4 KB 22.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4631_fsc.xml emd_4631_fsc.xml | 6.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_4631.png emd_4631.png | 38.9 KB | ||
| Masks |  emd_4631_msk_1.map emd_4631_msk_1.map | 12.9 MB |  Mask map Mask map | |
| Others |  emd_4631_additional.map.gz emd_4631_additional.map.gz emd_4631_additional_1.map.gz emd_4631_additional_1.map.gz | 1.6 MB 1.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4631 http://ftp.pdbj.org/pub/emdb/structures/EMD-4631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4631 | HTTPS FTP |
-Validation report
| Summary document |  emd_4631_validation.pdf.gz emd_4631_validation.pdf.gz | 255.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4631_full_validation.pdf.gz emd_4631_full_validation.pdf.gz | 254.6 KB | Display | |
| Data in XML |  emd_4631_validation.xml.gz emd_4631_validation.xml.gz | 8.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4631 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4631 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4631 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4631 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4631.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4631.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked dimer component of alpha-1 antitrypsin polymers isolated from explant liver tissue; subunits rotated ~90 degrees around dimer axis. Partially resolved third subunit evident. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.54 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4631_msk_1.map emd_4631_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
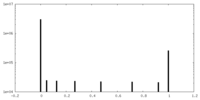
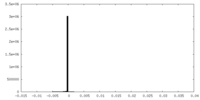
| Projections & Slices |
| ||||||||||||
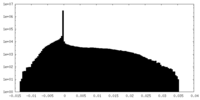
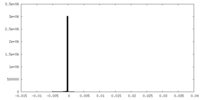
| Density Histograms |
-Additional map: Tightly masked dimer component of alpha-1 antitrypsin polymers...
| File | emd_4631_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tightly masked dimer component of alpha-1 antitrypsin polymers isolated from explant liver tissue; subunits rotated ~90 degrees around dimer axis. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Tightly masked dimer component of alpha-1 antitrypsin polymers...
| File | emd_4631_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tightly masked dimer component of alpha-1 antitrypsin polymers isolated from explant liver tissue; subunits rotated ~90 degrees around dimer axis. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Polymers of the severe deficiency Z variant of alpha-1 antitrypsi...
| Entire | Name: Polymers of the severe deficiency Z variant of alpha-1 antitrypsin labelled with 4B12 Fab fragments |
|---|---|
| Components |
|
-Supramolecule #1: Polymers of the severe deficiency Z variant of alpha-1 antitrypsi...
| Supramolecule | Name: Polymers of the severe deficiency Z variant of alpha-1 antitrypsin labelled with 4B12 Fab fragments type: tissue / ID: 1 / Parent: 0 / Macromolecule list: all Details: Polymers of alpha-1 antitrypsin were isolated from patient explant tissue and labelled with the Fab fragment of a non-conformationally selective monoclonal antibody (4B12). |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: Liver / Tissue: Explant tissue Homo sapiens (human) / Organ: Liver / Tissue: Explant tissue |
-Supramolecule #2: Complex between two 4B12 Fab fragments and a dimer component of p...
| Supramolecule | Name: Complex between two 4B12 Fab fragments and a dimer component of polymers of the severe deficiency Z alpha-1 antitrypsin variant type: complex / ID: 2 / Parent: 1 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: Liver Homo sapiens (human) / Organ: Liver |
| Molecular weight | Experimental: 200 KDa |
-Macromolecule #1: Alpha-1 antitrypsin
| Macromolecule | Name: Alpha-1 antitrypsin / type: protein_or_peptide / ID: 1 Details: 'Z' severe deficiency variant of alpha-1 antitrypsin Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: Liver / Tissue: Explant tissue Homo sapiens (human) / Organ: Liver / Tissue: Explant tissue |
| Sequence | String: EDPQGDAAQK TDTSHHDQDH PTFNKITPNL AEFAFS LYR QLAHQSNSTN IFFSPVSIAT AFAMLSLGTK ADTHDEILEG LNFNLTEIPE AQIHEGF QE LLRTLNQPDS QLQLTTGNGL FLSEGLKLVD KFLEDVKKLY HSEAFTVNFG DTEEAKKQ I NDYVEKGTQG ...String: EDPQGDAAQK TDTSHHDQDH PTFNKITPNL AEFAFS LYR QLAHQSNSTN IFFSPVSIAT AFAMLSLGTK ADTHDEILEG LNFNLTEIPE AQIHEGF QE LLRTLNQPDS QLQLTTGNGL FLSEGLKLVD KFLEDVKKLY HSEAFTVNFG DTEEAKKQ I NDYVEKGTQG KIVDLVKELD RDTVFALVNY IFFKGKWERP FEVKDTEEED FHVDQVTTV KVPMMKRLGM FNIQHCKKLS SWVLLMKYLG NATAIFFLPD EGKLQHLENE LTHDIITKFL ENEDRRSAS LHLPKLSITG TYDLKSVLGQ LGITKVFSNG ADLSGVTEEA PLKLSKAVHK A VLTIDKKG TEAAGAMFLE AIPMSIPPEV KFNKPFVFLM IEQNTKSPLF MGKVVNPTQK |
-Macromolecule #2: 4B12 Fab heavy chain
| Macromolecule | Name: 4B12 Fab heavy chain / type: protein_or_peptide / ID: 2 Details: Heavy chain of the Fab fragment of a non-conformationally-selective antibody (4B12) that binds to alpha-1-antitrypsin Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: QVKLEESGPE LVKPGASVKI SCKASGYSFI GYYMHWVKQS HVKSLEWIGR INPYNGATRY NQNFQDRATL TVDKSSSTAY MDFHSLTSED SAVYYCVRWP GDYWGQGTSV TVSSAKTTPP SVYPLAPGSA AQTNSMVTLG CLVKGYFPEP VTVTWNSGSL SSGVHTFPAV ...String: QVKLEESGPE LVKPGASVKI SCKASGYSFI GYYMHWVKQS HVKSLEWIGR INPYNGATRY NQNFQDRATL TVDKSSSTAY MDFHSLTSED SAVYYCVRWP GDYWGQGTSV TVSSAKTTPP SVYPLAPGSA AQTNSMVTLG CLVKGYFPEP VTVTWNSGSL SSGVHTFPAV LQSDLYTLSS SVTVPSSTWP SETVTCNVAH PASSTKVDKK IVPRDCTS |
-Macromolecule #3: 4B12 Fab light chain
| Macromolecule | Name: 4B12 Fab light chain / type: protein_or_peptide / ID: 3 Details: Light chain of the Fab fragment of a non-conformationally-selective antibody (4B12) that binds to alpha-1-antitrypsin Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: DIVMTQTPSS LSASLGGKVT ITCKASQDIN NYIAWYQLKP GKGPRQLIHY TSKLQPGIPS RFSGSGSGSD YSFSISNLEP EDIGTYYCLR YEDLWTFGGG TKLEIKRADA APTVSIFPPS SEQLTSGGAS VVCFLNNFYP KDINVKWKID GSERQNGVLN SWTDQDSKDS ...String: DIVMTQTPSS LSASLGGKVT ITCKASQDIN NYIAWYQLKP GKGPRQLIHY TSKLQPGIPS RFSGSGSGSD YSFSISNLEP EDIGTYYCLR YEDLWTFGGG TKLEIKRADA APTVSIFPPS SEQLTSGGAS VVCFLNNFYP KDINVKWKID GSERQNGVLN SWTDQDSKDS TYSMSSTLTL TKDEYERHNS YTCEATHKTS TSPIVKSFN |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl acetate Details: 3 microlitres of prepared sample was applied to glow-discharged carbon film on 300 mesh copper (CF300-Cu) grids. Sample was bound to the grid for 1 minute before blotting excess. 5 ...Details: 3 microlitres of prepared sample was applied to glow-discharged carbon film on 300 mesh copper (CF300-Cu) grids. Sample was bound to the grid for 1 minute before blotting excess. 5 microlitres of 2% w/v uranyl acetate was applied for 1 minute before blotting; this staining step was repeated once. | ||||||||||||
| Grid | Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||
| Details | The sample comprised a mixed population of unbranched polymers of varying lengths, labelled with the Fab fragment of the 4B12 antibody. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Digitization - Dimensions - Width: 5120 pixel / Digitization - Dimensions - Height: 3840 pixel / Digitization - Sampling interval: 6.4 µm / Digitization - Frames/image: 2-30 / Number grids imaged: 1 / Number real images: 100 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.009 µm / Calibrated defocus min: 0.888 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal magnification: 41470 |
| Sample stage | Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X