[English] 日本語
 Yorodumi
Yorodumi- EMDB-4577: Structure of fatty acid synthase complex with bound gamma subunit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4577 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of fatty acid synthase complex with bound gamma subunit from Saccharomyces cerevisiae at 2.8 angstrom | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Fatty acid synthase / Acyl carrier protein / Ketosynthase / Ketoreductase / Enoyl reductase / Dehydratase / Malonyl/palmitoyl transferase / Acetyl transferase / Phosphopantetheine transferase / transferase | |||||||||
| Function / homology |  Function and homology information Function and homology informationfatty-acyl-CoA synthase system / fatty-acyl-CoA synthase activity / fatty acid synthase complex / palmitoyltransferase activity / proteasome regulatory particle assembly / [acyl-carrier-protein] S-acetyltransferase / [acyl-carrier-protein] S-acetyltransferase activity / holo-[acyl-carrier-protein] synthase activity / enoyl-[acyl-carrier-protein] reductase (NADPH) activity / [acyl-carrier-protein] S-malonyltransferase ...fatty-acyl-CoA synthase system / fatty-acyl-CoA synthase activity / fatty acid synthase complex / palmitoyltransferase activity / proteasome regulatory particle assembly / [acyl-carrier-protein] S-acetyltransferase / [acyl-carrier-protein] S-acetyltransferase activity / holo-[acyl-carrier-protein] synthase activity / enoyl-[acyl-carrier-protein] reductase (NADPH) activity / [acyl-carrier-protein] S-malonyltransferase / [acyl-carrier-protein] S-malonyltransferase activity / 3-hydroxyacyl-[acyl-carrier-protein] dehydratase / (3R)-hydroxyacyl-[acyl-carrier-protein] dehydratase activity / beta-ketoacyl-[acyl-carrier-protein] synthase I / 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) activity / 3-oxoacyl-[acyl-carrier-protein] reductase / oleoyl-[acyl-carrier-protein] hydrolase / fatty acyl-[ACP] hydrolase activity / enoyl-[acyl-carrier-protein] reductase (NADH) / enoyl-[acyl-carrier-protein] reductase (NADH) activity / long-chain fatty acid biosynthetic process / fatty acid synthase activity / 3-oxoacyl-[acyl-carrier-protein] synthase activity / lipid droplet / enzyme activator activity / fatty acid biosynthetic process / protein-macromolecule adaptor activity / magnesium ion binding / mitochondrion / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
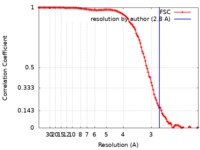
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Singh K / Graf B | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase. Authors: Kashish Singh / Benjamin Graf / Andreas Linden / Viktor Sautner / Henning Urlaub / Kai Tittmann / Holger Stark / Ashwin Chari /  Abstract: Fatty acid synthases (FASs) are central to metabolism but are also of biotechnological interest for the production of fine chemicals and biofuels from renewable resources. During fatty acid ...Fatty acid synthases (FASs) are central to metabolism but are also of biotechnological interest for the production of fine chemicals and biofuels from renewable resources. During fatty acid synthesis, the growing fatty acid chain is thought to be shuttled by the dynamic acyl carrier protein domain to several enzyme active sites. Here, we report the discovery of a γ subunit of the 2.6 megadalton α-βS. cerevisiae FAS, which is shown by high-resolution structures to stabilize a rotated FAS conformation and rearrange ACP domains from equatorial to axial positions. The γ subunit spans the length of the FAS inner cavity, impeding reductase activities of FAS, regulating NADPH turnover by kinetic hysteresis at the ketoreductase, and suppressing off-pathway reactions at the enoylreductase. The γ subunit delineates the functional compartment within FAS. As a scaffold, it may be exploited to incorporate natural and designed enzymatic activities that are not present in natural FAS. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4577.map.gz emd_4577.map.gz | 113 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4577-v30.xml emd-4577-v30.xml emd-4577.xml emd-4577.xml | 27.8 KB 27.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4577_fsc.xml emd_4577_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_4577.png emd_4577.png | 81.8 KB | ||
| Filedesc metadata |  emd-4577.cif.gz emd-4577.cif.gz | 9.3 KB | ||
| Others |  emd_4577_additional.map.gz emd_4577_additional.map.gz emd_4577_half_map_1.map.gz emd_4577_half_map_1.map.gz emd_4577_half_map_2.map.gz emd_4577_half_map_2.map.gz | 96.9 MB 97.6 MB 97.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4577 http://ftp.pdbj.org/pub/emdb/structures/EMD-4577 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4577 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4577 | HTTPS FTP |
-Validation report
| Summary document |  emd_4577_validation.pdf.gz emd_4577_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4577_full_validation.pdf.gz emd_4577_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_4577_validation.xml.gz emd_4577_validation.xml.gz | 18.6 KB | Display | |
| Data in CIF |  emd_4577_validation.cif.gz emd_4577_validation.cif.gz | 24.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4577 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4577 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4577 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4577 | HTTPS FTP |
-Related structure data
| Related structure data |  6ql5MC  4578C  6ql6C  6ql7C  6ql9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10454 (Title: Saccharomyces cerevisiae fatty acid synthase complex with bound gamma subunit EMPIAR-10454 (Title: Saccharomyces cerevisiae fatty acid synthase complex with bound gamma subunitData size: 329.3 Data #1: Aligned and doseweighted micrographs of yeast fatty acid synthase with bound gamma subunit [micrographs - single frame] Data #2: Stack of Polished particles (in relion) used for final reconstruction [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4577.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4577.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unsharpened map
| File | emd_4577_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
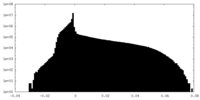
| Projections & Slices |
| ||||||||||||
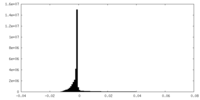
| Density Histograms |
-Half map: half map 2
| File | emd_4577_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
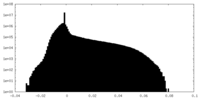
| Projections & Slices |
| ||||||||||||
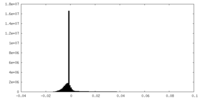
| Density Histograms |
-Half map: half map 1
| File | emd_4577_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Fatty acid synthase holoenzyme complex at 2.8 angstrom resolution
| Entire | Name: Fatty acid synthase holoenzyme complex at 2.8 angstrom resolution |
|---|---|
| Components |
|
-Supramolecule #1: Fatty acid synthase holoenzyme complex at 2.8 angstrom resolution
| Supramolecule | Name: Fatty acid synthase holoenzyme complex at 2.8 angstrom resolution type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Strain: BJ2168 (MATa prc1-407 prb1-1122 pep4-3 leu2 trp1 ura3-52 gal2 tma17::kanMX) |
| Molecular weight | Theoretical: 2.6 MDa |
-Macromolecule #1: Fatty acid synthase subunit alpha
| Macromolecule | Name: Fatty acid synthase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: fatty-acyl-CoA synthase system |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 207.184422 KDa |
| Sequence | String: MKPEVEQELA HILLTELLAY QFASPVRWIE TQDVFLKDFN TERVVEIGPS PTLAGMAQRT LKNKYESYDA ALSLHREILC YSKDAKEIY YTPDPSELAA KEEPAKEEAP APTPAASAPA PAAAAPAPVA AAAPAAAAAE IADEPVKASL LLHVLVAHKL K KSLDSIPM ...String: MKPEVEQELA HILLTELLAY QFASPVRWIE TQDVFLKDFN TERVVEIGPS PTLAGMAQRT LKNKYESYDA ALSLHREILC YSKDAKEIY YTPDPSELAA KEEPAKEEAP APTPAASAPA PAAAAPAPVA AAAPAAAAAE IADEPVKASL LLHVLVAHKL K KSLDSIPM SKTIKDLVGG KSTVQNEILG DLGKEFGTTP EKPEETPLEE LAETFQDTFS GALGKQSSSL LSRLISSKMP GG FTITVAR KYLQTRWGLP SGRQDGVLLV ALSNEPAARL GSEADAKAFL DSMAQKYASI VGVDLSSAAS ASGAAGAGAA AGA AMIDAG ALEEITKDHK VLARQQLQVL ARYLKMDLDN GERKFLKEKD TVAELQAQLD YLNAELGEFF VNGVATSFSR KKAR TFDSS WNWAKQSLLS LYFEIIHGVL KNVDREVVSE AINIMNRSND ALIKFMEYHI SNTDETKGEN YQLVKTLGEQ LIENC KQVL DVDPVYKDVA KPTGPKTAID KNGNITYSEE PREKVRKLSQ YVQEMALGGP ITKESQPTIE EDLTRVYKAI SAQADK QDI SSSTRVEFEK LYSDLMKFLE SSKEIDPSQT TQLAGMDVED ALDKDSTKEV ASLPNKSTIS KTVSSTIPRE TIPFLHL RK KTPAGDWKYD RQLSSLFLDG LEKAAFNGVT FKDKYVLITG AGKGSIGAEV LQGLLQGGAK VVVTTSRFSK QVTDYYQS I YAKYGAKGST LIVVPFNQGS KQDVEALIEF IYDTEKNGGL GWDLDAIIPF AAIPEQGIEL EHIDSKSEFA HRIMLTNIL RMMGCVKKQK SARGIETRPA QVILPMSPNH GTFGGDGMYS ESKLSLETLF NRWHSESWAN QLTVCGAIIG WTRGTGLMSA NNIIAEGIE KMGVRTFSQK EMAFNLLGLL TPEVVELCQK SPVMADLNGG LQFVPELKEF TAKLRKELVE TSEVRKAVSI E TALEHKVV NGNSADAAYA QVEIQPRANI QLDFPELKPY KQVKQIAPAE LEGLLDLERV IVVTGFAEVG PWGSARTRWE ME AFGEFSL EGCVEMAWIM GFISYHNGNL KGRPYTGWVD SKTKEPVDDK DVKAKYETSI LEHSGIRLIE PELFNGYNPE KKE MIQEVI VEEDLEPFEA SKETAEQFKH QHGDKVDIFE IPETGEYSVK LLKGATLYIP KALRFDRLVA GQIPTGWNAK TYGI SDDII SQVDPITLFV LVSVVEAFIA SGITDPYEMY KYVHVSEVGN CSGSGMGGVS ALRGMFKDRF KDEPVQNDIL QESFI NTMS AWVNMLLISS SGPIKTPVGA CATSVESVDI GVETILSGKA RICIVGGYDD FQEEGSFEFG NMKATSNTLE EFEHGR TPA EMSRPATTTR NGFMEAQGAG IQIIMQADLA LKMGVPIYGI VAMAATATDK IGRSVPAPGK GILTTAREHH SSVKYAS PN LNMKYRKRQL VTREAQIKDW VENELEALKL EAEEIPSEDQ NEFLLERTRE IHNEAESQLR AAQQQWGNDF YKRDPRIA P LRGALATYGL TIDDLGVASF HGTSTKANDK NESATINEMM KHLGRSEGNP VIGVFQKFLT GHPKGAAGAW MMNGALQIL NSGIIPGNRN ADNVDKILEQ FEYVLYPSKT LKTDGVRAVS ITSFGFGQKG GQAIVVHPDY LYGAITEDRY NEYVAKVSAR EKSAYKFFH NGMIYNKLFV SKEHAPYTDE LEEDVYLDPL ARVSKDKKSG SLTFNSKNIQ SKDSYINANT IETAKMIENM T KEKVSNGG VGVDVELITS INVENDTFIE RNFTPQEIEY CSAQPSVQSS FAGTWSAKEA VFKSLGVKSL GGGAALKDIE IV RVNKNAP AVELHGNAKK AAEEAGVTDV KVSISHDDLQ AVAVAVSTKK UniProtKB: Fatty acid synthase subunit alpha |
-Macromolecule #2: Fatty acid synthase subunit beta
| Macromolecule | Name: Fatty acid synthase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO / EC number: fatty-acyl-CoA synthase system |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 227.785141 KDa |
| Sequence | String: STRPLTLSHG SLEHVLLVPT ASFFIASQLQ EQFNKILPEP TEGFAADDEP TTPAELVGKF LGYVSSLVEP SKVGQFDQVL NLCLTEFEN CYLEGNDIHA LAAKLLQEND TTLVKTKELI KNYITARIMA KRPFDKKSNS ALFRAVGEGN AQLVAIFGGQ G NTDDYFEE ...String: STRPLTLSHG SLEHVLLVPT ASFFIASQLQ EQFNKILPEP TEGFAADDEP TTPAELVGKF LGYVSSLVEP SKVGQFDQVL NLCLTEFEN CYLEGNDIHA LAAKLLQEND TTLVKTKELI KNYITARIMA KRPFDKKSNS ALFRAVGEGN AQLVAIFGGQ G NTDDYFEE LRDLYQTYHV LVGDLIKFSA ETLSELIRTT LDAEKVFTQG LNILEWLENP SNTPDKDYLL SIPISCPLIG VI QLAHYVV TAKLLGFTPG ELRSYLKGAT GHSQGLVTAV AIAETDSWES FFVSVRKAIT VLFFIGVRCY EAYPNTSLPP SIL EDSLEN NEGVPSPMLS ISNLTQEQVQ DYVNKTNSHL PAGKQVEISL VNGAKNLVVS GPPQSLYGLN LTLRKAKAPS GLDQ SRIPF SERKLKFSNR FLPVASPFHS HLLVPASDLI NKDLVKNNVS FNAKDIQIPV YDTFDGSDLR VLSGSISERI VDCII RLPV KWETTTQFKA THILDFGPGG ASGLGVLTHR NKDGTGVRVI VAGTLDINPD DDYGFKQEIF DVTSNGLKKN PNWLEE YHP KLIKNKSGKI FVETKFSKLI GRPPLLVPGM TPCTVSPDFV AATTNAGYTI ELAGGGYFSA AGMTAAIDSV VSQIEKG ST FGINLIYVNP FMLQWGIPLI KELRSKGYPI QFLTIGAGVP SLEVASEYIE TLGLKYLGLK PGSIDAISQV INIAKAHP N FPIALQWTGG RGGGHHSFED AHTPMLQMYS KIRRHPNIML IFGSGFGSAD DTYPYLTGEW STKFDYPPMP FDGFLFGSR VMIAKEVKTS PDAKKCIAAC TGVPDDKWEQ TYKKPTGGIV TVRSEMGEPI HKIATRGVML WKEFDETIFN LPKNKLVPTL EAKRDYIIS RLNADFQKPW FATVNGQARD LATMTYEEVA KRLVELMFIR STNSWFDVTW RTFTGDFLRR VEERFTKSKT L SLIQSYSL LDKPDEAIEK VFNAYPAARE QFLNAQDIDH FLSMCQNPMQ KPVPFVPVLD RRFEIFFKKD SLWQSEHLEA VV DQDVQRT CILHGPVAAQ FTKVIDEPIK SIMDGIHDGH IKKLLHQYYG DDESKIPAVE YFGGESPVDV QSDSEDSAVF KAT SSTDEE SWFKALAGSE INWRHASFLC SFITQDKMFV SNPIRKVFKP SQGMVVEISN GNTSSKTVVT LSEPVQGELK PTVI LKLLK ENIIQMEMIE NRTMDGKPVS LPLLYNFNPD NGFAPISEVM EDRNQRIKEM YWKLWIDEPF NLDFDPRDVI KGKDF EITA KEVYDFTHAV GNNCEDFVSR PDRTMLAPMD FAIVVGWRAI IKAIFPNTVD GDLLKLVHLS NGYKMIPGAK PLQVGD VVS TTAVIESVVN QPTGKIVDVV GTLSRNGKPV MEVTSSFFYR GNYTDFENTF QKTVEPVYQM HIKTSKDIAV LRSKEWF QL DDEDFDLLNK TLTFETETEV TFKNANIFSS VKCFGPIKVE LPTKETVEIG IVDYEAGASH GNPVVDFLKR NGSTLEQK V NLENPIPIAV LDSYTPSTNE PYARVSGDLN PIHVSRHFAS YANLPGTITH GMFSSASVRA LIENWAADSV SSRVRGYTC QFVDMVLPNT ALKTSIQHVG MINGRKLIKF ETRNEDDVVV LTGEAEIEQP VTTFVFTGQG SQEQGMGMDL YKTSKAAQDV WNRADNHFK DTYGFSILDI VINNPVNLTI HFGGEKGKRI RENYSAMIFE TIVDGKLKTE KIFKEINEHS TSYTFRSEKG L LSATQFTQ PALTLMEKAA FEDLKSKGLI PADATFAGHS LGEYAALASL ADVMSIESLV EVVFYRGMTM QVAVPRDELG RS NYGMIAI NPGRVAASFS QEALQYVVER VGKRTGWLVE IVNYNVENQQ YVAAGDLRAL DTVTNVLNFI KLQKIDIIEL QKS LSLEEV EGHLFEIIDE ASKKSAVKPR PLKLERGFAC IPLVGISVPF HSTYLMNGVK PFKSFLKKNI IKENVKVARL AGKY IPNLT AKPFQVTKEY FQDVYDLTGS EPIKEIIDNW EKYEQ UniProtKB: Fatty acid synthase subunit beta |
-Macromolecule #3: Translation machinery-associated protein 17
| Macromolecule | Name: Translation machinery-associated protein 17 / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.558117 KDa |
| Sequence | String: SAGGIRRPIQ IEEFKTAISG MSDMELAQIK TEIENSINHL QRSNARLGKY IAKLEGADDR LEADDSDDLE NIDSGDLALY KDSVRENEI VLNNYNERVD ALEQETVYRK TGHGKSKHEV EAKDNTNKGP DVDMDNSNVD VVTPNSIFI UniProtKB: Translation machinery-associated protein 17 |
-Macromolecule #4: 4'-PHOSPHOPANTETHEINE
| Macromolecule | Name: 4'-PHOSPHOPANTETHEINE / type: ligand / ID: 4 / Number of copies: 6 / Formula: PNS |
|---|---|
| Molecular weight | Theoretical: 358.348 Da |
| Chemical component information |  ChemComp-PNS: |
-Macromolecule #5: FLAVIN MONONUCLEOTIDE
| Macromolecule | Name: FLAVIN MONONUCLEOTIDE / type: ligand / ID: 5 / Number of copies: 6 / Formula: FMN |
|---|---|
| Molecular weight | Theoretical: 456.344 Da |
| Chemical component information |  ChemComp-FMN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
| ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number real images: 5441 / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 132000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)