+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1623 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cerulenin-inhibited type I yeast Fatty Acid Synthase | |||||||||
 Map data Map data | This is a cryo-EM map of cerulenin inhibited yeast FAS filtered to 5.9A resolution. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | fatty acid synthase / type I fungal FAS mechanism / yeast / fatty acid synthesis. | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.9 Å | |||||||||
 Authors Authors | Gipson P / Mills DJ / Wouts R / Grininger M / Vonck J / Kuehlbrandt W | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2010 Journal: Proc Natl Acad Sci U S A / Year: 2010Title: Direct structural insight into the substrate-shuttling mechanism of yeast fatty acid synthase by electron cryomicroscopy. Authors: Preeti Gipson / Deryck J Mills / Remco Wouts / Martin Grininger / Janet Vonck / Werner Kühlbrandt /  Abstract: Yeast fatty acid synthase (FAS) is a 2.6-MDa barrel-shaped multienzyme complex, which carries out cyclic synthesis of fatty acids. By electron cryomicroscopy of single particles we obtained a three- ...Yeast fatty acid synthase (FAS) is a 2.6-MDa barrel-shaped multienzyme complex, which carries out cyclic synthesis of fatty acids. By electron cryomicroscopy of single particles we obtained a three-dimensional map of yeast FAS at 5.9-A resolution. Compared to the crystal structures of fungal FAS, the EM map reveals major differences and new features that indicate a considerably different arrangement of the complex in solution compared to the crystal structures, as well as a high degree of variance inside the barrel. Distinct density regions in the reaction chambers next to each of the catalytic domains fitted the substrate-binding acyl carrier protein (ACP) domain. In each case, this resulted in the expected distance of approximately 18 A from the ACP substrate-binding site to the active site of the catalytic domains. The multiple, partially occupied positions of the ACP within the reaction chamber provide direct structural insight into the substrate-shuttling mechanism of fatty acid synthesis in this large cellular machine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1623.map.gz emd_1623.map.gz | 92.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1623-v30.xml emd-1623-v30.xml emd-1623.xml emd-1623.xml | 8.3 KB 8.3 KB | Display Display |  EMDB header EMDB header |
| Images |  FAS-1000dpi.tiff FAS-1000dpi.tiff FAS-emdb.tif FAS-emdb.tif | 732.9 KB 396.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1623 http://ftp.pdbj.org/pub/emdb/structures/EMD-1623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1623 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1623.map.gz / Format: CCP4 / Size: 96.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1623.map.gz / Format: CCP4 / Size: 96.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a cryo-EM map of cerulenin inhibited yeast FAS filtered to 5.9A resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.14 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
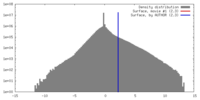
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cerulenin-inhibited Yeast FAS
| Entire | Name: Cerulenin-inhibited Yeast FAS |
|---|---|
| Components |
|
-Supramolecule #1000: Cerulenin-inhibited Yeast FAS
| Supramolecule | Name: Cerulenin-inhibited Yeast FAS / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 2.6 MDa |
-Macromolecule #1: Type I yeast FAS
| Macromolecule | Name: Type I yeast FAS / type: protein_or_peptide / ID: 1 / Name.synonym: yeast FAS / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: Plunger |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 1.19 µm / Number real images: 150 / Average electron dose: 25 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: each partilce |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D3 (2x3 fold dihedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 5.9 Å / Resolution method: OTHER / Software - Name: EMAN1 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Automatic rigid body fits |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)