[English] 日本語
 Yorodumi
Yorodumi- EMDB-45218: Human DNA polymerase theta helicase domain in microhomology annea... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human DNA polymerase theta helicase domain in microhomology annealed state 2, dimer form | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA repair / helicase / ATPase / TRANSFERASE-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsingle-stranded DNA endodeoxyribonuclease activity / HDR through MMEJ (alt-NHEJ) / single-stranded DNA helicase activity / double-strand break repair via alternative nonhomologous end joining / replication fork processing / site of DNA damage / 5'-deoxyribose-5-phosphate lyase activity / somatic hypermutation of immunoglobulin genes / negative regulation of double-strand break repair via homologous recombination / error-prone translesion synthesis ...single-stranded DNA endodeoxyribonuclease activity / HDR through MMEJ (alt-NHEJ) / single-stranded DNA helicase activity / double-strand break repair via alternative nonhomologous end joining / replication fork processing / site of DNA damage / 5'-deoxyribose-5-phosphate lyase activity / somatic hypermutation of immunoglobulin genes / negative regulation of double-strand break repair via homologous recombination / error-prone translesion synthesis / DNA helicase activity / base-excision repair / protein homooligomerization / RNA-directed DNA polymerase / RNA-directed DNA polymerase activity / double-strand break repair / site of double-strand break / DNA helicase / damaged DNA binding / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA repair / DNA damage response / chromatin binding / Golgi apparatus / magnesium ion binding / ATP hydrolysis activity / nucleoplasm / ATP binding / identical protein binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
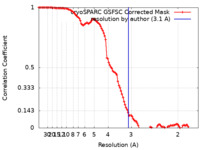
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Ito F / Li Z / Chen XS | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2024 Journal: bioRxiv / Year: 2024Title: Structural Basis for Polθ-Helicase DNA Binding and Microhomology-Mediated End-Joining. Authors: Fumiaki Ito / Ziyuan Li / Leonid Minakhin / Htet A Khant / Richard T Pomerantz / Xiaojiang S Chen /  Abstract: DNA double-strand breaks (DSBs) present a critical threat to genomic integrity, often precipitating genomic instability and oncogenesis. Repair of DSBs predominantly occurs through homologous ...DNA double-strand breaks (DSBs) present a critical threat to genomic integrity, often precipitating genomic instability and oncogenesis. Repair of DSBs predominantly occurs through homologous recombination (HR) and non-homologous end joining (NHEJ). In HR-deficient cells, DNA polymerase theta (Polθ) becomes critical for DSB repair via microhomology-mediated end joining (MMEJ), also termed theta-mediated end joining (TMEJ). Thus, Polθ is synthetically lethal with BRCA1/2 and other HR factors, underscoring its potential as a therapeutic target in HR-deficient cancers. However, the molecular mechanisms governing Polθ-mediated MMEJ remain poorly understood. Here we present a series of cryo-electron microscopy structures of the Polθ helicase domain (Polθ-hel) in complex with DNA containing 3'-overhang. The structures reveal the sequential conformations adopted by Polθ-hel during the critical phases of DNA binding, microhomology searching, and microhomology annealing. The stepwise conformational changes within the Polθ-hel subdomains and its functional dimeric state are pivotal for aligning the 3'-overhangs, facilitating the microhomology search and subsequent annealing necessary for DSB repair via MMEJ. Our findings illustrate the essential molecular switches within Polθ-hel that orchestrate the MMEJ process in DSB repair, laying the groundwork for the development of targeted therapies against the Polθ-hel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45218.map.gz emd_45218.map.gz | 64.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45218-v30.xml emd-45218-v30.xml emd-45218.xml emd-45218.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45218_fsc.xml emd_45218_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_45218.png emd_45218.png | 146 KB | ||
| Filedesc metadata |  emd-45218.cif.gz emd-45218.cif.gz | 6.1 KB | ||
| Others |  emd_45218_half_map_1.map.gz emd_45218_half_map_1.map.gz emd_45218_half_map_2.map.gz emd_45218_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45218 http://ftp.pdbj.org/pub/emdb/structures/EMD-45218 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45218 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45218 | HTTPS FTP |
-Related structure data
| Related structure data |  9c5qMC  8w0aC  9asjC  9askC  9aslC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45218.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45218.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_45218_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
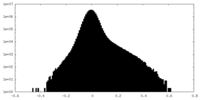
| Projections & Slices |
| ||||||||||||
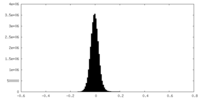
| Density Histograms |
-Half map: #2
| File | emd_45218_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
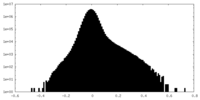
| Projections & Slices |
| ||||||||||||
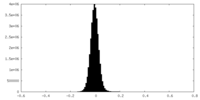
| Density Histograms |
- Sample components
Sample components
-Entire : Human DNA polymerase theta helicase domain in microhomology annea...
| Entire | Name: Human DNA polymerase theta helicase domain in microhomology annealed state 2, dimer form |
|---|---|
| Components |
|
-Supramolecule #1: Human DNA polymerase theta helicase domain in microhomology annea...
| Supramolecule | Name: Human DNA polymerase theta helicase domain in microhomology annealed state 2, dimer form type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 253 KDa |
-Macromolecule #1: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*CP*CP*CP*GP*GP*G)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*CP*CP*CP*GP*GP*G)-3') type: dna / ID: 1 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4.243748 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DC)(DC) (DC)(DG)(DG)(DG) |
-Macromolecule #2: DNA polymerase theta
| Macromolecule | Name: DNA polymerase theta / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 92.089797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KLLLANWGLP KAVLEKYHSF GVKKMFEWQA ECLLLGQVLE GKNLVYSAPT SAGKTLVAEL LILKRVLEMR KKALFILPFV SVAKEKKYY LQSLFQEVGI KVDGYMGSTS PSRHFSSLDI AVCTIERANG LINRLIEENK MDLLGMVVVD ELHMLGDSHR G YLLELLLT ...String: KLLLANWGLP KAVLEKYHSF GVKKMFEWQA ECLLLGQVLE GKNLVYSAPT SAGKTLVAEL LILKRVLEMR KKALFILPFV SVAKEKKYY LQSLFQEVGI KVDGYMGSTS PSRHFSSLDI AVCTIERANG LINRLIEENK MDLLGMVVVD ELHMLGDSHR G YLLELLLT KICYITRKSA SCQADLASSL SNAVQIVGMS ATLPNLELVA SWLNAELYHT DFRPVPLLES VKVGNSIYDS SM KLVREFE PMLQVKGDED HVVSLCYETI CDNHSVLLFC PSKKWCEKLA DIIAREFYNL HHQAEGLVKP SECPPVILEQ KEL LEVMDQ LRRLPSGLDS VLQKTVPWGV AFHHAGLTFE ERDIIEGAFR QGLIRVLAAT STLSSGVNLP ARRVIIRTPI FGGR PLDIL TYKQMVGRAG RKGVDTVGES ILICKNSEKS KGIALLQGSL KPVRSCLQRR EGEEVTGSMI RAILEIIVGG VASTS QDMH TYAACTFLAA SMKEGKQGIQ RNQESVQLGA IEACVMWLLE NEFIQSTEAS DGTEGKVYHP THLGSATLSS SLSPAD TLD IFADLQRAMK GFVLENDLHI LYLVTPMFED WTTIDWYRFF CLWEKLPTSM KRVAELVGVE EGFLARCVAA AAAAAAA AQ HRQMAIHKRF FTSLVLLDLI SEVPLREINQ KYGCNRGQIQ SLQQSAAVYA GMITVFSNRL GWHNMELLLS QFQKRLTF G IQRELCDLVR VSLLNAQRAR VLYASGFHTV ADLARANIVE VEVILKNAVP FKSARKAVDE EEEAVEERRN MRTIWVTGR KGLTEREAAA LIVEEARMIL QQDL UniProtKB: DNA polymerase theta |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 10005 / Average exposure time: 3.5 sec. / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X