+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-4450 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Localized reconstruction of tail-spike of bacteriophage P68 | ||||||||||||
 マップデータ マップデータ | Localized reconstruction of tail-spike of bacteriophage P68 | ||||||||||||
 試料 試料 |
| ||||||||||||
| 生物種 |   Staphylococcus phage P68 (ファージ) Staphylococcus phage P68 (ファージ) | ||||||||||||
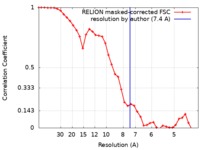
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 7.4 Å | ||||||||||||
 データ登録者 データ登録者 | Hrebik D / Skubnik K / Fuzik T / Plevka P | ||||||||||||
| 資金援助 |  チェコ, 3件 チェコ, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: Sci Adv / 年: 2019 ジャーナル: Sci Adv / 年: 2019タイトル: Structure and genome ejection mechanism of phage P68. 著者: Dominik Hrebík / Dana Štveráková / Karel Škubník / Tibor Füzik / Roman Pantůček / Pavel Plevka /  要旨: Phages infecting can be used as therapeutics against antibiotic-resistant bacterial infections. However, there is limited information about the mechanism of genome delivery of phages that infect ...Phages infecting can be used as therapeutics against antibiotic-resistant bacterial infections. However, there is limited information about the mechanism of genome delivery of phages that infect Gram-positive bacteria. Here, we present the structures of native phage P68, genome ejection intermediate, and empty particle. The P68 head contains 72 subunits of inner core protein, 15 of which bind to and alter the structure of adjacent major capsid proteins and thus specify attachment sites for head fibers. Unlike in the previously studied phages, the head fibers of P68 enable its virion to position itself at the cell surface for genome delivery. The unique interaction of one end of P68 DNA with one of the 12 portal protein subunits is disrupted before the genome ejection. The inner core proteins are released together with the DNA and enable the translocation of phage genome across the bacterial membrane into the cytoplasm. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_4450.map.gz emd_4450.map.gz | 2.8 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-4450-v30.xml emd-4450-v30.xml emd-4450.xml emd-4450.xml | 14.7 KB 14.7 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_4450_fsc.xml emd_4450_fsc.xml | 3.7 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_4450.png emd_4450.png | 69 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4450 http://ftp.pdbj.org/pub/emdb/structures/EMD-4450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4450 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_4450_validation.pdf.gz emd_4450_validation.pdf.gz | 234.9 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_4450_full_validation.pdf.gz emd_4450_full_validation.pdf.gz | 234.1 KB | 表示 | |
| XML形式データ |  emd_4450_validation.xml.gz emd_4450_validation.xml.gz | 7.3 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4450 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4450 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4450 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4450 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  4435C  4436C  4437C  4438C  4440C  4442C  4449C  4451C  4453C  4454C  4455C  4456C  4457C  4458C  4459C  6iabC  6iacC  6iatC  6iawC  6ib1C  6q3gC C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_4450.map.gz / 形式: CCP4 / 大きさ: 3.8 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_4450.map.gz / 形式: CCP4 / 大きさ: 3.8 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Localized reconstruction of tail-spike of bacteriophage P68 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 2.126 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Staphylococcus phage P68
| 全体 | 名称:   Staphylococcus phage P68 (ファージ) Staphylococcus phage P68 (ファージ) |
|---|---|
| 要素 |
|
-超分子 #1: Staphylococcus phage P68
| 超分子 | 名称: Staphylococcus phage P68 / タイプ: virus / ID: 1 / 親要素: 0 / 含まれる分子: all / NCBI-ID: 204090 / 生物種: Staphylococcus phage P68 / ウイルスタイプ: VIRION / ウイルス・単離状態: STRAIN / ウイルス・エンベロープ: No / ウイルス・中空状態: No |
|---|---|
| 宿主 | 生物種:  |
| 分子量 | 理論値: 6.9 KDa |
| ウイルス殻 | Shell ID: 1 / 名称: Capsid / 直径: 480.0 Å / T番号(三角分割数): 4 |
-分子 #1: Tail-spike of bacteriophage P68
| 分子 | 名称: Tail-spike of bacteriophage P68 / タイプ: protein_or_peptide / ID: 1 / 光学異性体: LEVO / EC番号: lysozyme |
|---|---|
| 配列 | 文字列: MNDQEKIDKF THSYINDDFG LTIDQLVPKV KGYGRFNVWL GGNESKIRQV LKAVKEIGVS PTLFAVYEKN EGFSSGLGW LNHTSARGDY LTDAKFIARK LVSQSKQAGQ PSWYDAGNIV HFVPQDVQRK GNADFAKNMK A GTIGRAYI PLTAAATWAA YYPLGLKASY ...文字列: MNDQEKIDKF THSYINDDFG LTIDQLVPKV KGYGRFNVWL GGNESKIRQV LKAVKEIGVS PTLFAVYEKN EGFSSGLGW LNHTSARGDY LTDAKFIARK LVSQSKQAGQ PSWYDAGNIV HFVPQDVQRK GNADFAKNMK A GTIGRAYI PLTAAATWAA YYPLGLKASY NKVQNYGNPF LDGANTILAW GGKLDGKGGS PSDSSDSGSS GD SGSSLLA LAKQAMQELL KKIQDALQWD VHSIGSDKFF SNDYFTLEKT FNNTYHIKMT IGLLDSLKKL IDS VQVDSG SSSSNPTDDD GDHKPISGKS VKPNGKSGRV IGGNWTYAQL PEKYKKAIGV PLFKKEYLYK PGNI FPQTG NAGQCTELTW AYMSQLHGKR QPTDDGQITN GQRVWYVYKK LGAKTTHNPT VGYGFSSKPP YLQAT AYGI GHTGVVVAVF EDGSFLVANY NVPPYVAPSR VVLYTLINGV PNNAGDNIVF FSGIA |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 8 構成要素:
| ||||||||||||
| グリッド | モデル: Quantifoil R2/1 / 材質: COPPER / メッシュ: 200 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 雰囲気: NITROGEN | ||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 293 K / 装置: FEI VITROBOT MARK IV / 詳細: blot time 2s; blot force -2; 3.6 ul of sample. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: FEI FALCON II (4k x 4k) 検出モード: INTEGRATING / デジタル化 - サイズ - 横: 4096 pixel / デジタル化 - サイズ - 縦: 4096 pixel / デジタル化 - 画像ごとのフレーム数: 1-7 / 撮影したグリッド数: 2 / 実像数: 2891 / 平均露光時間: 1.0 sec. / 平均電子線量: 21.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 70.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 0.003 µm / 最小 デフォーカス(公称値): 0.001 µm / 倍率(公称値): 75000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー