[English] 日本語
 Yorodumi
Yorodumi- EMDB-4131: Structure of the mammalian ribosomal termination complex with eRF... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4131 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the mammalian ribosomal termination complex with eRF1 and eRF3. | ||||||||||||
 Map data Map data | Postprocess, sharpened map. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Translation / Elongation / Ribosome | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationtranslation termination factor activity / translation release factor complex / cytoplasmic translational termination / regulation of translational termination / translation release factor activity, codon specific / protein methylation / ribosomal subunit / translation release factor activity / sequence-specific mRNA binding / peptidyl-tRNA hydrolase activity ...translation termination factor activity / translation release factor complex / cytoplasmic translational termination / regulation of translational termination / translation release factor activity, codon specific / protein methylation / ribosomal subunit / translation release factor activity / sequence-specific mRNA binding / peptidyl-tRNA hydrolase activity / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / Protein hydroxylation / Eukaryotic Translation Termination / ubiquitin ligase inhibitor activity / positive regulation of signal transduction by p53 class mediator / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / 90S preribosome / phagocytic cup / translational termination / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / rough endoplasmic reticulum / translation regulator activity / ribosomal small subunit export from nucleus / gastrulation / MDM2/MDM4 family protein binding / cytosolic ribosome / class I DNA-(apurinic or apyrimidinic site) endonuclease activity / DNA-(apurinic or apyrimidinic site) lyase / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / positive regulation of apoptotic signaling pathway / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / small-subunit processome / G1/S transition of mitotic cell cycle / Regulation of expression of SLITs and ROBOs / spindle / rRNA processing / antimicrobial humoral immune response mediated by antimicrobial peptide / rhythmic process / positive regulation of canonical Wnt signaling pathway / regulation of translation / large ribosomal subunit / ribosome binding / ribosomal small subunit biogenesis / ribosomal small subunit assembly / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / defense response to Gram-negative bacterium / perikaryon / cytosolic large ribosomal subunit / killing of cells of another organism / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / cytoplasmic translation / cell differentiation / tRNA binding / mitochondrial inner membrane / rRNA binding / postsynaptic density / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / cell division / DNA repair / GTPase activity / mRNA binding / apoptotic process / synapse / dendrite / centrosome / GTP binding / nucleolus / perinuclear region of cytoplasm / endoplasmic reticulum / Golgi apparatus / DNA binding / RNA binding / zinc ion binding / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.65 Å | ||||||||||||
 Authors Authors | Shao S / Murray J | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2016 Journal: Cell / Year: 2016Title: Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes. Authors: Sichen Shao / Jason Murray / Alan Brown / Jack Taunton / V Ramakrishnan / Ramanujan S Hegde /   Abstract: In eukaryotes, accurate protein synthesis relies on a family of translational GTPases that pair with specific decoding factors to decipher the mRNA code on ribosomes. We present structures of the ...In eukaryotes, accurate protein synthesis relies on a family of translational GTPases that pair with specific decoding factors to decipher the mRNA code on ribosomes. We present structures of the mammalian ribosome engaged with decoding factor⋅GTPase complexes representing intermediates of translation elongation (aminoacyl-tRNA⋅eEF1A), termination (eRF1⋅eRF3), and ribosome rescue (Pelota⋅Hbs1l). Comparative analyses reveal that each decoding factor exploits the plasticity of the ribosomal decoding center to differentially remodel ribosomal proteins and rRNA. This leads to varying degrees of large-scale ribosome movements and implies distinct mechanisms for communicating information from the decoding center to each GTPase. Additional structural snapshots of the translation termination pathway reveal the conformational changes that choreograph the accommodation of decoding factors into the peptidyl transferase center. Our results provide a structural framework for how different states of the mammalian ribosome are selectively recognized by the appropriate decoding factor⋅GTPase complex to ensure translational fidelity. #1:  Journal: To Be Published Journal: To Be PublishedTitle: Decoding mammalian ribosome-mRNA states by translational GTPase complexes Authors: Shao S / Murray J / Brown A / Taunton J / Ramakrishnan V / Hegde RS | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4131.map.gz emd_4131.map.gz | 13 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4131-v30.xml emd-4131-v30.xml emd-4131.xml emd-4131.xml | 108.3 KB 108.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4131_fsc.xml emd_4131_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_4131.png emd_4131.png | 185.8 KB | ||
| Filedesc metadata |  emd-4131.cif.gz emd-4131.cif.gz | 21.5 KB | ||
| Others |  emd_4131_half_map_1.map.gz emd_4131_half_map_1.map.gz emd_4131_half_map_2.map.gz emd_4131_half_map_2.map.gz | 245.2 MB 245.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4131 http://ftp.pdbj.org/pub/emdb/structures/EMD-4131 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4131 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4131 | HTTPS FTP |
-Validation report
| Summary document |  emd_4131_validation.pdf.gz emd_4131_validation.pdf.gz | 800.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4131_full_validation.pdf.gz emd_4131_full_validation.pdf.gz | 800.5 KB | Display | |
| Data in XML |  emd_4131_validation.xml.gz emd_4131_validation.xml.gz | 22.8 KB | Display | |
| Data in CIF |  emd_4131_validation.cif.gz emd_4131_validation.cif.gz | 30.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4131 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4131 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4131 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4131 | HTTPS FTP |
-Related structure data
| Related structure data |  5lztMC  4129C  4130C  4132C  4133C  4134C  4135C  4136C  4137C  5lzsC  5lzuC  5lzvC  5lzwC  5lzxC  5lzyC  5lzzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4131.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4131.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocess, sharpened map. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
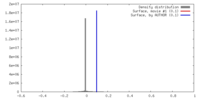
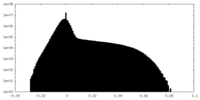
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 1.
| File | emd_4131_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2.
| File | emd_4131_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Affinity-purified 80S ribosome-nascent chain complex reconstitute...
+Supramolecule #1: Affinity-purified 80S ribosome-nascent chain complex reconstitute...
+Macromolecule #1: uL2
+Macromolecule #2: uL3
+Macromolecule #3: uL4
+Macromolecule #4: uL18
+Macromolecule #5: eL6
+Macromolecule #6: uL30
+Macromolecule #7: eL8
+Macromolecule #8: uL6
+Macromolecule #9: uL16
+Macromolecule #10: uL5
+Macromolecule #11: eL13
+Macromolecule #12: eL14
+Macromolecule #13: eL15
+Macromolecule #14: uL13
+Macromolecule #15: uL22
+Macromolecule #16: eL18
+Macromolecule #17: eL19
+Macromolecule #18: eL20
+Macromolecule #19: eL21
+Macromolecule #20: eL22
+Macromolecule #21: uL14
+Macromolecule #22: eL24
+Macromolecule #23: uL23
+Macromolecule #24: uL24
+Macromolecule #25: eL27
+Macromolecule #26: uL15
+Macromolecule #27: eL29
+Macromolecule #28: eL30
+Macromolecule #29: eL31
+Macromolecule #30: eL32
+Macromolecule #31: eL33
+Macromolecule #32: eL34
+Macromolecule #33: uL29
+Macromolecule #34: eL36
+Macromolecule #35: eL37
+Macromolecule #36: eL38
+Macromolecule #37: eL39
+Macromolecule #38: eL40
+Macromolecule #39: eL41
+Macromolecule #40: eL42
+Macromolecule #41: eL43
+Macromolecule #42: eL28
+Macromolecule #43: uL10
+Macromolecule #44: uL11
+Macromolecule #45: Nascent chain
+Macromolecule #52: uS2
+Macromolecule #53: eS1
+Macromolecule #54: uS5
+Macromolecule #55: uS3
+Macromolecule #56: eS4
+Macromolecule #57: uS7
+Macromolecule #58: eS6
+Macromolecule #59: eS7
+Macromolecule #60: eS8
+Macromolecule #61: uS4
+Macromolecule #62: eS10
+Macromolecule #63: uS17
+Macromolecule #64: eS12
+Macromolecule #65: uS15
+Macromolecule #66: uS11
+Macromolecule #67: uS19
+Macromolecule #68: uS9
+Macromolecule #69: eS17
+Macromolecule #70: uS13
+Macromolecule #71: eS19
+Macromolecule #72: uS10
+Macromolecule #73: eS21
+Macromolecule #74: uS8
+Macromolecule #75: uS12
+Macromolecule #76: eS24
+Macromolecule #77: eS25
+Macromolecule #78: eS26
+Macromolecule #79: eS27
+Macromolecule #80: eS28
+Macromolecule #81: uS14
+Macromolecule #82: eS30
+Macromolecule #83: eS31
+Macromolecule #84: RACK1
+Macromolecule #86: eRF1
+Macromolecule #87: eRF3a
+Macromolecule #46: P-site tRNA
+Macromolecule #47: E-site tRNA
+Macromolecule #48: 28S ribosomal RNA
+Macromolecule #49: 5S ribosomal RNA
+Macromolecule #50: 5.8S ribosomal RNA
+Macromolecule #51: 18S ribosomal RNA
+Macromolecule #85: mRNA (UGA stop codon)
+Macromolecule #88: MAGNESIUM ION
+Macromolecule #89: ZINC ION
+Macromolecule #90: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5 / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III Details: 3 ul aliquots were applied to the grid and incubated for 30 s, before blotting for 3s to remove excess solution.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Number real images: 2611 / Average exposure time: 1.0 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 104478 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Overall B value: 63.3 / Target criteria: FSCaverage |
|---|---|
| Output model |  PDB-5lzt: |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)