[English] 日本語
 Yorodumi
Yorodumi- EMDB-40887: Raw consensus map of TRPM2 chanzyme (E1114A) in the presence of M... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Raw consensus map of TRPM2 chanzyme (E1114A) in the presence of Magnesium and ADP-ribose, open state | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | TRPM2 Chanzyme / Channel-enzyme / MEMBRANE PROTEIN | |||||||||||||||
| Biological species |  Salpingoeca rosetta (eukaryote) Salpingoeca rosetta (eukaryote) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.87 Å | |||||||||||||||
 Authors Authors | Huang Y / Kumar S / Lu W / Du J | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution. Authors: Yihe Huang / Sushant Kumar / Junuk Lee / Wei Lü / Juan Du /  Abstract: Channel enzymes represent a class of ion channels with enzymatic activity directly or indirectly linked to their channel function. We investigated a TRPM2 chanzyme from choanoflagellates that ...Channel enzymes represent a class of ion channels with enzymatic activity directly or indirectly linked to their channel function. We investigated a TRPM2 chanzyme from choanoflagellates that integrates two seemingly incompatible functions into a single peptide: a channel module activated by ADP-ribose with high open probability and an enzyme module (NUDT9-H domain) consuming ADP-ribose at a remarkably slow rate. Using time-resolved cryogenic-electron microscopy, we captured a complete series of structural snapshots of gating and catalytic cycles, revealing the coupling mechanism between channel gating and enzymatic activity. The slow kinetics of the NUDT9-H enzyme module confers a self-regulatory mechanism: ADPR binding triggers NUDT9-H tetramerization, promoting channel opening, while subsequent hydrolysis reduces local ADPR, inducing channel closure. We further demonstrated how the NUDT9-H domain has evolved from a structurally semi-independent ADP-ribose hydrolase module in early species to a fully integrated component of a gating ring essential for channel activation in advanced species. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40887.map.gz emd_40887.map.gz | 252.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40887-v30.xml emd-40887-v30.xml emd-40887.xml emd-40887.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40887.png emd_40887.png | 132.6 KB | ||
| Filedesc metadata |  emd-40887.cif.gz emd-40887.cif.gz | 5.7 KB | ||
| Others |  emd_40887_additional_1.map.gz emd_40887_additional_1.map.gz emd_40887_half_map_1.map.gz emd_40887_half_map_1.map.gz emd_40887_half_map_2.map.gz emd_40887_half_map_2.map.gz | 30.4 MB 253.2 MB 253.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40887 http://ftp.pdbj.org/pub/emdb/structures/EMD-40887 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40887 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40887 | HTTPS FTP |
-Validation report
| Summary document |  emd_40887_validation.pdf.gz emd_40887_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40887_full_validation.pdf.gz emd_40887_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_40887_validation.xml.gz emd_40887_validation.xml.gz | 16.8 KB | Display | |
| Data in CIF |  emd_40887_validation.cif.gz emd_40887_validation.cif.gz | 20 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40887 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40887 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40887 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40887 | HTTPS FTP |
-Related structure data
| Related structure data |  8sr7C  8sr8C  8sr9C  8sraC  8srbC  8srcC  8srdC  8sreC  8srfC  8srgC  8srhC  8sriC  8srjC  8srkC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40887.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40887.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||
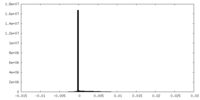
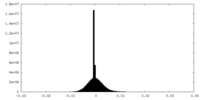
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_40887_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
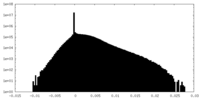
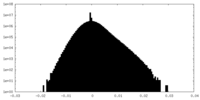
| Density Histograms |
-Half map: #1
| File | emd_40887_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
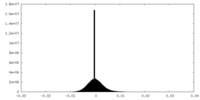
| Density Histograms |
-Half map: #2
| File | emd_40887_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
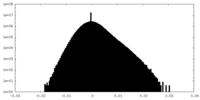
| Density Histograms |
- Sample components
Sample components
-Entire : TRPM2 chanzyme (E1114A) incubated with Magnesium and ADP-ribose f...
| Entire | Name: TRPM2 chanzyme (E1114A) incubated with Magnesium and ADP-ribose for 5s; ADP-ribose intact; open state |
|---|---|
| Components |
|
-Supramolecule #1: TRPM2 chanzyme (E1114A) incubated with Magnesium and ADP-ribose f...
| Supramolecule | Name: TRPM2 chanzyme (E1114A) incubated with Magnesium and ADP-ribose for 5s; ADP-ribose intact; open state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Salpingoeca rosetta (eukaryote) Salpingoeca rosetta (eukaryote) |
-Macromolecule #1: TRPM2 chanzyme (E1114A) incubated with Magnesium and ADP-ribose f...
| Macromolecule | Name: TRPM2 chanzyme (E1114A) incubated with Magnesium and ADP-ribose for 5s; ADP-ribose intact; open state. type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salpingoeca rosetta (eukaryote) Salpingoeca rosetta (eukaryote) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQRARPGELV EVIMFRPTGK ARVSNLDESM AMEFTDLRTR AMSSAAMIRQ SVAAKTLLIE NEDGKGSTRM EVQDFMKRFH MHASEDDKT GSPSTAWGTL RFPTKEATAP YLRLSVNDDP EDALLFVKAM LAQKYGETYD RPSLILSVTG GARNFTLPPR L ETAIAKGL ...String: MQRARPGELV EVIMFRPTGK ARVSNLDESM AMEFTDLRTR AMSSAAMIRQ SVAAKTLLIE NEDGKGSTRM EVQDFMKRFH MHASEDDKT GSPSTAWGTL RFPTKEATAP YLRLSVNDDP EDALLFVKAM LAQKYGETYD RPSLILSVTG GARNFTLPPR L ETAIAKGL RLAAQRTNAW VVTGGTNTGV MKLTGQIMEA LSKTQSHFIP PTIGIATYGV IIGGDDMTRG EPPKIGLEYE MH KKDPPKT TPLDDNHNLF LLVDDGSTNK FGKEIKFRAA FENAAGQAFA APVVTIVVQG GPGTLGTALQ AVRQGTPIVV VDG SGLAAD VLAYAYNFMH NPLTRFKSYT IDDLRQKVAQ TFNPKSSQQL TNLLDSALEC VQDPNLVVVY SLQESGIDEF DDCI LKAIF SSQGKLGNKL KQAMYFDQLD VAKRALSEAS KNGQHNEIAA CINDNLMAAM MHNKPHFVEL YLGFDAKIYE LKPSE EVAK TNITALDELP SFALAIEELY KREAKKPHSH VQRLVSLSNT DVLGRHYRVS TQRGDGTTRR IGRDLANTRA YNVLRM DQI FARLVSKDFS VNRDFTIYDS KYDKVPGIQF RRTAQASHML FLWAICLDRF RMARHFWLIG DQSIINALVA SRILERL ST HRALQGPHLA EERAKMQHNA KKFEELAVGV LGECHGSDSH MASEMLHSKN DMFNKKNAIN IAYDAKSLAF LSHPATQS V INADWYGHLK SVTSFWAVLF AFFFPFFVLP FINFSEDHAE QQVEAPRDFF TDAPRSSHSA NSTTSGAHRL RRKFAKFYS APYTRFISDL LSHFVLCVVT SYFVLDKLED TISAIEWILL VWFVALLLEE LRQMIFCDGI AEYISDTWNR LDLIMITLFF VGFFTHASD PSNQDSKVVS KGIHAFLVVV LWLRFMRYYA LSKNLGPKLI MMMEMMKDVS TFVFLLLIFL IGYGVAAQSL L SPDEDFSS RTFIGVLFRP YFQIYGELFL DDLNSEANCL GDTPFTECSR ETVRMVPFFL AVYILGSNVL LVNLLIAMFN DT YMKVQEA AEDLWRKQNY ELCAEYKDRP FLPAPFILLA HVHMLFMRLL RLCGVHTQEH EKIQDDETKR KITTFEALNT DKF LRRWER ERQEMLEARV KMTNDNVVQA MGMMDQLLEH MISFRFSLDQ QATKIKQEIR DDGLPSTEPT GLVSRTPSQP INRL NSAVA VHGHTAEAAE WYVPPEEYPK SGGVKRYLID ASMVPLSIMC PSYDPVEYTH PSVAAQPVWA DPADPRKIKF NVKDE VNGK VVDRTSCHPS GISIDSNTGR PINPWGRTGM TGRGLLGKWG VNQAADTVVT RWKRSPDGSI LERDGKKVLE FVAIQR QDN KMWAIPGGFV DNGEDVALTS GREFMEEALG MGTSADLMSA ESKDSLAALF SSGTIVARIY CEDPRNTDNA WVETTCV NF HDESGRHAAR LKLQGGDDAE HARWMMVHGG LNLFASHRTL LQHVTSALNA YF(APR)(MG)(MG)(MG)(MG)(MG) (CLR) (CLR)(CLR)(APR) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 0.02 sec. / Average electron dose: 49.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.9000000000000001 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.87 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 8279 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)