[English] 日本語
 Yorodumi
Yorodumi- EMDB-37243: Structure of the human ATP synthase bound to bedaquiline (membran... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human ATP synthase bound to bedaquiline (membrane domain) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ATP synthase / Human / cryo-EM / Membrane protein | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationFormation of ATP by chemiosmotic coupling / Cristae formation / ATP biosynthetic process / Mitochondrial protein import / mitochondrial proton-transporting ATP synthase complex assembly / : / oxidative phosphorylation / response to copper ion / proton-transporting ATP synthase complex / : ...Formation of ATP by chemiosmotic coupling / Cristae formation / ATP biosynthetic process / Mitochondrial protein import / mitochondrial proton-transporting ATP synthase complex assembly / : / oxidative phosphorylation / response to copper ion / proton-transporting ATP synthase complex / : / : / : / proton-transporting ATP synthase complex, coupling factor F(o) / proton motive force-driven ATP synthesis / proton motive force-driven mitochondrial ATP synthesis / proton transmembrane transporter activity / response to hyperoxia / Mitochondrial protein degradation / substantia nigra development / proton-transporting ATP synthase activity, rotational mechanism / aerobic respiration / proton transmembrane transport / nuclear membrane / mitochondrial inner membrane / hydrolase activity / mitochondrial matrix / lipid binding / mitochondrion / RNA binding / membrane / nucleus Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.13 Å | ||||||||||||
 Authors Authors | Lai Y / Zhang Y / Gong H | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587. Authors: Yuying Zhang / Yuezheng Lai / Shan Zhou / Ting Ran / Yue Zhang / Ziqing Zhao / Ziyan Feng / Long Yu / Jinxu Xu / Kun Shi / Jianyun Wang / Yu Pang / Liang Li / Hongming Chen / Luke W Guddat / ...Authors: Yuying Zhang / Yuezheng Lai / Shan Zhou / Ting Ran / Yue Zhang / Ziqing Zhao / Ziyan Feng / Long Yu / Jinxu Xu / Kun Shi / Jianyun Wang / Yu Pang / Liang Li / Hongming Chen / Luke W Guddat / Yan Gao / Fengjiang Liu / Zihe Rao / Hongri Gong /   Abstract: Bedaquiline (BDQ), a first-in-class diarylquinoline anti-tuberculosis drug, and its analogue, TBAJ-587, prevent the growth and proliferation of Mycobacterium tuberculosis by inhibiting ATP synthase. ...Bedaquiline (BDQ), a first-in-class diarylquinoline anti-tuberculosis drug, and its analogue, TBAJ-587, prevent the growth and proliferation of Mycobacterium tuberculosis by inhibiting ATP synthase. However, BDQ also inhibits human ATP synthase. At present, how these compounds interact with either M. tuberculosis ATP synthase or human ATP synthase is unclear. Here we present cryogenic electron microscopy structures of M. tuberculosis ATP synthase with and without BDQ and TBAJ-587 bound, and human ATP synthase bound to BDQ. The two inhibitors interact with subunit a and the c-ring at the leading site, c-only sites and lagging site in M. tuberculosis ATP synthase, showing that BDQ and TBAJ-587 have similar modes of action. The quinolinyl and dimethylamino units of the compounds make extensive contacts with the protein. The structure of human ATP synthase in complex with BDQ reveals that the BDQ-binding site is similar to that observed for the leading site in M. tuberculosis ATP synthase, and that the quinolinyl unit also interacts extensively with the human enzyme. This study will improve researchers' understanding of the similarities and differences between human ATP synthase and M. tuberculosis ATP synthase in terms of the mode of BDQ binding, and will allow the rational design of novel diarylquinolines as anti-tuberculosis drugs. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37243.map.gz emd_37243.map.gz | 483.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37243-v30.xml emd-37243-v30.xml emd-37243.xml emd-37243.xml | 25.1 KB 25.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37243.png emd_37243.png | 50.1 KB | ||
| Filedesc metadata |  emd-37243.cif.gz emd-37243.cif.gz | 6.5 KB | ||
| Others |  emd_37243_half_map_1.map.gz emd_37243_half_map_1.map.gz emd_37243_half_map_2.map.gz emd_37243_half_map_2.map.gz | 475.6 MB 475.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37243 http://ftp.pdbj.org/pub/emdb/structures/EMD-37243 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37243 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37243 | HTTPS FTP |
-Validation report
| Summary document |  emd_37243_validation.pdf.gz emd_37243_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37243_full_validation.pdf.gz emd_37243_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_37243_validation.xml.gz emd_37243_validation.xml.gz | 18.9 KB | Display | |
| Data in CIF |  emd_37243_validation.cif.gz emd_37243_validation.cif.gz | 22.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37243 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37243 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37243 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37243 | HTTPS FTP |
-Related structure data
| Related structure data |  8khfMC  8j0sC  8j0tC  8j57C  8j58C  8jr0C  8jr1C  8ki3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37243.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37243.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.73 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_37243_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_37243_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : human ATP synthase
+Supramolecule #1: human ATP synthase
+Macromolecule #1: ATP synthase F(0) complex subunit C1, mitochondrial
+Macromolecule #2: ATP synthase subunit gamma, mitochondrial
+Macromolecule #3: ATP synthase subunit delta, mitochondrial
+Macromolecule #4: ATP synthase subunit epsilon, mitochondrial
+Macromolecule #5: ATP synthase F(0) complex subunit B1, mitochondrial
+Macromolecule #6: ATP synthase subunit d, mitochondrial
+Macromolecule #7: ATP synthase subunit a
+Macromolecule #8: ATP synthase subunit ATP5MJ, mitochondrial
+Macromolecule #9: ATP synthase protein 8
+Macromolecule #10: ATP synthase subunit f, mitochondrial
+Macromolecule #11: ATP synthase subunit g, mitochondrial
+Macromolecule #12: ATP synthase subunit e, mitochondrial
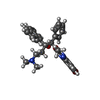
+Macromolecule #13: Bedaquiline
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.13 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 84037 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)