+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3506 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryoEM structure of bacterial holo-translocon | ||||||||||||
 Map data Map data | map of cross-linked HTL | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | holotranslocon / membrane protein insertion machinery / chaperone / protein secretion | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotein insertion into membrane from inner side / membrane insertase activity / cell envelope Sec protein transport complex / protein transport by the Sec complex / intracellular protein transmembrane transport / protein-transporting ATPase activity / SRP-dependent cotranslational protein targeting to membrane, translocation / signal sequence binding / protein insertion into membrane / protein transmembrane transporter activity ...protein insertion into membrane from inner side / membrane insertase activity / cell envelope Sec protein transport complex / protein transport by the Sec complex / intracellular protein transmembrane transport / protein-transporting ATPase activity / SRP-dependent cotranslational protein targeting to membrane, translocation / signal sequence binding / protein insertion into membrane / protein transmembrane transporter activity / protein secretion / protein targeting / intracellular protein transport / protein transport / protein folding / protein-containing complex assembly / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
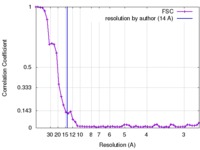
| Method | single particle reconstruction / cryo EM / Resolution: 14.0 Å | ||||||||||||
 Authors Authors | Schaffitzel C / Botte M / Karuppasamy M / Papai G / Schultz P | ||||||||||||
| Funding support |  Germany, Germany,  France, France,  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2016 Journal: Sci Rep / Year: 2016Title: A central cavity within the holo-translocon suggests a mechanism for membrane protein insertion. Authors: Mathieu Botte / Nathan R Zaccai / Jelger Lycklama À Nijeholt / Remy Martin / Kèvin Knoops / Gabor Papai / Juan Zou / Aurélien Deniaud / Manikandan Karuppasamy / Qiyang Jiang / Abhishek ...Authors: Mathieu Botte / Nathan R Zaccai / Jelger Lycklama À Nijeholt / Remy Martin / Kèvin Knoops / Gabor Papai / Juan Zou / Aurélien Deniaud / Manikandan Karuppasamy / Qiyang Jiang / Abhishek Singha Roy / Klaus Schulten / Patrick Schultz / Juri Rappsilber / Giuseppe Zaccai / Imre Berger / Ian Collinson / Christiane Schaffitzel /     Abstract: The conserved SecYEG protein-conducting channel and the accessory proteins SecDF-YajC and YidC constitute the bacterial holo-translocon (HTL), capable of protein-secretion and membrane-protein ...The conserved SecYEG protein-conducting channel and the accessory proteins SecDF-YajC and YidC constitute the bacterial holo-translocon (HTL), capable of protein-secretion and membrane-protein insertion. By employing an integrative approach combining small-angle neutron scattering (SANS), low-resolution electron microscopy and biophysical analyses we determined the arrangement of the proteins and lipids within the super-complex. The results guided the placement of X-ray structures of individual HTL components and allowed the proposal of a model of the functional translocon. Their arrangement around a central lipid-containing pool conveys an unexpected, but compelling mechanism for membrane-protein insertion. The periplasmic domains of YidC and SecD are poised at the protein-channel exit-site of SecY, presumably to aid the emergence of translocating polypeptides. The SecY lateral gate for membrane-insertion is adjacent to the membrane 'insertase' YidC. Absolute-scale SANS employing a novel contrast-match-point analysis revealed a dynamic complex adopting open and compact configurations around an adaptable central lipid-filled chamber, wherein polytopic membrane-proteins could fold, sheltered from aggregation and proteolysis. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3506.map.gz emd_3506.map.gz | 9.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3506-v30.xml emd-3506-v30.xml emd-3506.xml emd-3506.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3506_fsc.xml emd_3506_fsc.xml | 5.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_3506.png emd_3506.png | 143.1 KB | ||
| Filedesc metadata |  emd-3506.cif.gz emd-3506.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3506 http://ftp.pdbj.org/pub/emdb/structures/EMD-3506 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3506 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3506 | HTTPS FTP |
-Validation report
| Summary document |  emd_3506_validation.pdf.gz emd_3506_validation.pdf.gz | 224.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3506_full_validation.pdf.gz emd_3506_full_validation.pdf.gz | 223.8 KB | Display | |
| Data in XML |  emd_3506_validation.xml.gz emd_3506_validation.xml.gz | 8.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3506 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3506 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3506 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3506 | HTTPS FTP |
-Related structure data
| Related structure data |  5mg3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3506.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3506.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map of cross-linked HTL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : bacterial holo-translocon (HTL)
| Entire | Name: bacterial holo-translocon (HTL) |
|---|---|
| Components |
|
-Supramolecule #1: bacterial holo-translocon (HTL)
| Supramolecule | Name: bacterial holo-translocon (HTL) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Membrane Protein Complex consisting of SecYEG-SecDFYajC-YidC |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: Protein translocase subunit SecY
| Macromolecule | Name: Protein translocase subunit SecY / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.410523 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VWNCERITIS HRKQTMAKQP GLDFQSAKGG LGELKRRLLF VIGALIVFRI GSFIPIPGID AAVLAKLLEQ QRGTIIEMFN MFSGGALSR ASIFALGIMP YISASIIIQL LTVVHPTLAE IKKEGESGRR KISQYTRYGT LVLAIFQSIG IATGLPNMPG M QGLVINPG ...String: VWNCERITIS HRKQTMAKQP GLDFQSAKGG LGELKRRLLF VIGALIVFRI GSFIPIPGID AAVLAKLLEQ QRGTIIEMFN MFSGGALSR ASIFALGIMP YISASIIIQL LTVVHPTLAE IKKEGESGRR KISQYTRYGT LVLAIFQSIG IATGLPNMPG M QGLVINPG FAFYFTAVVS LVTGTMFLMW LGEQITERGI GNGISIIIFA GIVAGLPPAI AHTIEQARQG DLHFLVLLLV AV LVFAVTF FVVFVERGQR RIVVNYAKRQ QGRRVYAAQS THLPLKVNMA GVIPAIFASS IILFPATIAS WFGGGTGWNW LTT ISLYLQ PGQPLYVLLY ASAIIFFCFF YTALVFNPRE TADNLKKSGA FVPGIRPGEQ TAKYIDKVMT RLTLVGALYI TFIC LIPEF MRDAMKVPFY FGGTSLLIVV VVIMDFMAQV QTLMMSSQYE SALKKANLKG YGR UniProtKB: Protein translocase subunit SecY |
-Macromolecule #2: Protein translocase subunit SecE
| Macromolecule | Name: Protein translocase subunit SecE / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.248021 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHDDD DKAMGANTEA QGSGRGLEAM KWVVVVALLL VAIVGNYLYR DIMLPLRALA VVILIAAAGG VALLTTKGKA TVAFAREAR TEVRKVIWPT RQETLHTTLI VAAVTAVMSL ILWGLDGILV RLVSFITGLR F UniProtKB: Protein translocase subunit SecE |
-Macromolecule #3: Protein-export membrane protein SecG
| Macromolecule | Name: Protein-export membrane protein SecG / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.326448 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VGTGWYSGSP GILYHWPEVL RIQELIMYEA LLVVFLIVAI GLVGLIMLQQ GKGADMGASF GAGASATLFG SSGSGNFMTR MTALLATLF FIISLVLGNI NSNKTNKGSE WENLSAPAKT EQTQPAAPAK PTSDIPN UniProtKB: Protein-export membrane protein SecG |
-Macromolecule #4: Protein translocase subunit SecD
| Macromolecule | Name: Protein translocase subunit SecD / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 67.687984 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHMLN RYPLWKYVML IVVIVIGLLY ALPNLFGEDP AVQITGARGV AASEQTLIQV QKTLQEEKIT AKSVALEEGA ILARFDSTD TQLRAREALM GVMGDKYVVA LNLAPATPRW LAAIHAEPMK LGLDLRGGVH FLMEVDMDTV LGKLQEQNID S LRSDLREK ...String: MHHHHHHMLN RYPLWKYVML IVVIVIGLLY ALPNLFGEDP AVQITGARGV AASEQTLIQV QKTLQEEKIT AKSVALEEGA ILARFDSTD TQLRAREALM GVMGDKYVVA LNLAPATPRW LAAIHAEPMK LGLDLRGGVH FLMEVDMDTV LGKLQEQNID S LRSDLREK GIPYTTVRKE NNYGLSITFR DAKARDEAIA YLSKRHPDLV ISSQGSNQLR AVMSDARLSE AREYAVQQNI NI LRNRVNQ LGVAEPVVQR QGADRIVVEL PGIQDTARAK EILGATATLE FRLVNTNVDQ AAAASGRVPG DSEVKQTREG QPV VLYKRV ILTGDHITDS TSSQDEYNQP QVNISLDSAG GNIMSNFTKD NIGKPMATLF VEYKDSGKKD ANGRAVLVKQ EEVI NIANI QSRLGNSFRI TGINNPNEAR QLSLLLRAGA LIAPIQIVEE RTIGPTLGMQ NIEQGLEACL AGLLVSILFM IIFYK KFGL IATSALIANL ILIVGIMSLL PGATLSMPGI AGIVLTLAVA VDANVLINER IKEELSNGRT VQQAIDEGYR GAFSSI FDA NITTLIKVII LYAVGTGAIK GFAITTGIGV ATSMFTAIVG TRAIVNLLYG GKRVKKLSI UniProtKB: Protein translocase subunit SecD |
-Macromolecule #5: Protein translocase subunit SecF
| Macromolecule | Name: Protein translocase subunit SecF / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.41325 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAQEYTVEQL NHGRKVYDFM RWDYWAFGIS GLLLIAAIVI MGVRGFNWGL DFTGGTVIEI TLEKPAEIDV MRDALQKAGF EEPMLQNFG SSHDIMVRMP PAEGETGGQV LGSQVLKVIN ESTNQNAAVK RIEFVGPSVG ADLAQTGAMA LMAALLSILV Y VGFRFEWR ...String: MAQEYTVEQL NHGRKVYDFM RWDYWAFGIS GLLLIAAIVI MGVRGFNWGL DFTGGTVIEI TLEKPAEIDV MRDALQKAGF EEPMLQNFG SSHDIMVRMP PAEGETGGQV LGSQVLKVIN ESTNQNAAVK RIEFVGPSVG ADLAQTGAMA LMAALLSILV Y VGFRFEWR LAAGVVIALA HDVIITLGIL SLFHIEIDLT IVASLMSVIG YSLNDSIVVS DRIRENFRKI RRGTPYEIFN VS LTQTLHR TLITSGTTLM VILMLYLFGG PVLEGFSLTM LIGVSIGTAS SIYVASALAL KLGMKREHML QQKVEKEGAD QPS ILP UniProtKB: Protein translocase subunit SecF |
-Macromolecule #6: Membrane protein insertase YidC
| Macromolecule | Name: Membrane protein insertase YidC / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 62.658582 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDPSSRDSQR NLLVIALLFV SFMIWQAWEQ DKNPQPQAQQ TTQTTTTAAG SAADQGVPAS GQGKLISVKT DVLDLTINTR GGDVEQALL PAYPKELNST QPFQLLETSP QFIYQAQSGL TGRDGPDNPA NGPRPLYNVE KDAYVLAEGQ NELQVPMTYT D AAGNTFTK ...String: MDPSSRDSQR NLLVIALLFV SFMIWQAWEQ DKNPQPQAQQ TTQTTTTAAG SAADQGVPAS GQGKLISVKT DVLDLTINTR GGDVEQALL PAYPKELNST QPFQLLETSP QFIYQAQSGL TGRDGPDNPA NGPRPLYNVE KDAYVLAEGQ NELQVPMTYT D AAGNTFTK TFVLKRGDYA VNVNYNVQNA GEKPLEISSF GQLKQSITLP PHLDTGSSNF ALHTFRGAAY STPDAAYAAY AF DTIADNE NLNISSKGGW VAMLQQYFAT AWIPHNDGTN NFYTANLGNG IAAIGYKSQP VLVQPGQTGA MNSTLWVGPE IQD KMAAVA PHLDLTVDYG WLWFISQPLF KLLKWIHSFV GNWGFSIIII TFIVRGIMYP LTKAQYTSMA KMRMLQPKIQ AMRE RLGDD KQRISQEMMA LYKAEKVNPL GGCFPLLIQM PIFLALYYML MGSVELRQAP FALWIHDLSA QDPYYILPIL MGVTM FFIQ KMSPTTVTDP MQQKIMTFMP VIFTVFFLWF PSGLVLYYIV SNLVTIIQQQ LIYRGLEKRG LHSREKKKSH HHHHH UniProtKB: Membrane protein insertase YidC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 5 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: FEI FALCON I (4k x 4k) / Average electron dose: 10.0 e/Å2 |
| Electron beam | Acceleration voltage: 100 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)