[English] 日本語
 Yorodumi
Yorodumi- EMDB-31183: In situ structure of transcriptional enzyme complex and capsid sh... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31183 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ structure of transcriptional enzyme complex and capsid shell protein of mammalian reovirus at initiation state | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationviral inner capsid / host cytoskeleton / 7-methylguanosine mRNA capping / viral genome replication /  viral capsid / viral nucleocapsid / host cell cytoplasm / viral capsid / viral nucleocapsid / host cell cytoplasm /  RNA-directed RNA polymerase / RNA-directed RNA polymerase /  RNA-dependent RNA polymerase activity / structural molecule activity / RNA-dependent RNA polymerase activity / structural molecule activity /  RNA binding RNA bindingSimilarity search - Function | ||||||||||||||||||
| Biological species |   Mammalian orthoreovirus 3 / Mammalian orthoreovirus 3 /   Mammalian orthoreovirus 3 Dearing Mammalian orthoreovirus 3 Dearing | ||||||||||||||||||
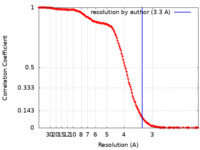
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.3 Å cryo EM / Resolution: 3.3 Å | ||||||||||||||||||
 Authors Authors | Zhou ZH / Pan M | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Asymmetric reconstruction of mammalian reovirus reveals interactions among RNA, transcriptional factor µ2 and capsid proteins. Authors: Muchen Pan / Ana L Alvarez-Cabrera / Joon S Kang / Lihua Wang / Chunhai Fan / Z Hong Zhou /   Abstract: Mammalian reovirus (MRV) is the prototypical member of genus Orthoreovirus of family Reoviridae. However, lacking high-resolution structures of its RNA polymerase cofactor μ2 and infectious ...Mammalian reovirus (MRV) is the prototypical member of genus Orthoreovirus of family Reoviridae. However, lacking high-resolution structures of its RNA polymerase cofactor μ2 and infectious particle, limits understanding of molecular interactions among proteins and RNA, and their contributions to virion assembly and RNA transcription. Here, we report the 3.3 Å-resolution asymmetric reconstruction of transcribing MRV and in situ atomic models of its capsid proteins, the asymmetrically attached RNA-dependent RNA polymerase (RdRp) λ3, and RdRp-bound nucleoside triphosphatase μ2 with a unique RNA-binding domain. We reveal molecular interactions among virion proteins and genomic and messenger RNA. Polymerase complexes in three Spinoreovirinae subfamily members are organized with different pseudo-D symmetries to engage their highly diversified genomes. The above interactions and those between symmetry-mismatched receptor-binding σ1 trimers and RNA-capping λ2 pentamers balance competing needs of capsid assembly, external protein removal, and allosteric triggering of endogenous RNA transcription, before, during and after infection, respectively. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31183.map.gz emd_31183.map.gz | 191.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31183-v30.xml emd-31183-v30.xml emd-31183.xml emd-31183.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31183_fsc.xml emd_31183_fsc.xml | 18.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_31183.png emd_31183.png | 147.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31183 http://ftp.pdbj.org/pub/emdb/structures/EMD-31183 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31183 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31183 | HTTPS FTP |
-Related structure data
| Related structure data |  7elhMC  7ellC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31183.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31183.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mammalian orthoreovirus 3 Dearing
| Entire | Name:   Mammalian orthoreovirus 3 Dearing Mammalian orthoreovirus 3 Dearing |
|---|---|
| Components |
|
-Supramolecule #1: Mammalian orthoreovirus 3 Dearing
| Supramolecule | Name: Mammalian orthoreovirus 3 Dearing / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3, #6 / NCBI-ID: 10886 / Sci species name: Mammalian orthoreovirus 3 Dearing / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Minor core protein mu2
| Macromolecule | Name: Minor core protein mu2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mammalian orthoreovirus 3 Mammalian orthoreovirus 3 |
| Molecular weight | Theoretical: 83.331289 KDa |
| Sequence | String: MAYIAVPAVV DSRSSEAIGL LESFGVDAGA DANDVSYQDH DYVLDQLQYM LDGYEAGDVI DALVHKNWLH HSVYCLLPPK SQLLEYWKS NPSAIPDNVD RRLRKRLMLK KDLRKDDEYN QLARAFKISD VYAPLISSTT SPMTMIQNLN QGEIVYTTTD R VIGARILL ...String: MAYIAVPAVV DSRSSEAIGL LESFGVDAGA DANDVSYQDH DYVLDQLQYM LDGYEAGDVI DALVHKNWLH HSVYCLLPPK SQLLEYWKS NPSAIPDNVD RRLRKRLMLK KDLRKDDEYN QLARAFKISD VYAPLISSTT SPMTMIQNLN QGEIVYTTTD R VIGARILL YAPRKYYAST LSFTMTKCII PFGKEVGRVP HSRFNVGTFP SIATPKCFVM SGVDIESIPN EFIKLFYQRV KS VHANILN DISPQIVSDM INRKRLRVHT PSDRRAAQLM HLPYHVKRGA SHVDVYKVDV VDMLFEVVDV ADGLRNVSRK LTM HTVPVC ILEMLGIEIA DYCIRQEDGM LTDWFLLLTM LSDGLTDRRT HCQYLINPSS VPPDVILNIS ITGFINRHTI DVMP DIYDF VKPIGAVLPK GSFKSTIMRV LDSISILGIQ IMPRAHVVDS DEVGEQMEPT FEQAVMEIYK GIAGVDSLDD LIKWV LNSD LIPHDDRLGQ LFQAFLPLAK DLLAPMARKF YDNSMSEGRL LTFAHADSEL LNANYFGHLL RLKIPYITEV NLMIRK NRE GGELFQLVLS YLYKMYATSA QPKWFGSLLR LLICPWLHME KLIGEADPAS TSAEIGWHIP REQLMQDGWC GCEDGFI PY VSIRAPRLVI EELMEKNWGQ YHAQVIVTDQ LVVGEPRRVS AKAVIKGNHL PVKLVSRFAC FTLTAKYEMR LSCGHSTG R GAAYSARLAF RSDLA |
-Macromolecule #2: RNA-directed RNA polymerase
| Macromolecule | Name: RNA-directed RNA polymerase / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number:  RNA-directed RNA polymerase RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:   Mammalian orthoreovirus 3 Mammalian orthoreovirus 3 |
| Molecular weight | Theoretical: 142.449016 KDa |
| Sequence | String: MSSMILTQFG PFIESISGIT DQSNDVFEDA AKAFSMFTRS DVYKALDEIP FSDDAMLPIP PTIYTKPSHD SYYYIDALNR VRRKTYQGP DDVYVPNCSI VELLEPHETL TSYGRLSEAI ENRAKDGDSQ ARIATTYGRI AESQARQIKA PLEKFVLALL V AEAGGSLY ...String: MSSMILTQFG PFIESISGIT DQSNDVFEDA AKAFSMFTRS DVYKALDEIP FSDDAMLPIP PTIYTKPSHD SYYYIDALNR VRRKTYQGP DDVYVPNCSI VELLEPHETL TSYGRLSEAI ENRAKDGDSQ ARIATTYGRI AESQARQIKA PLEKFVLALL V AEAGGSLY DPVLQKYDEI PDLSHNCPLW CFREICRHIS GPLPDRAPYL YLSAGVFWLM SPRMTSAIPP LLSDLVNLAI LQ QTAGLDP SLVKLGVQIC LHAAASSSYA WFILKTKSIF PQNTLHSMYE SLEGGYCPNL EWLEPRSDYK FMYMGVMPLS AKY ARSAPS NDKKARELGE KYGLSSVVGE LRKRTKTYVK HDFASVRYIR DAMACTSGIF LVRTPTETVL QEYTQSPEIK VPIP QKDWT GPIGEIRILK DTTSSIARYL YRTWYLAAAR MAAQPRTWDP LFQAIMRSQY VTARGGSGAA LRESLYAINV SLPDF KGLP VKAATKIFQA AQLANLPFSH TSVAILADTS MGLRNQVQRR PRSIMPLNVP QQQVSAPHTL TADYINYHMN LSTTSG SAV IEKVIPLGVY ASSPPNQSIN IDISACDASI TWDFFLSVIM AAIHEGVASS SIGKPFMGVP ASIVNDESVV GVRAARP IS GMQNMIQHLS KLYKRGFSYR VNDSFSPGND FTHMTTTFPS GSTATSTEHT ANNSTMMETF LTVWGPEHTD DPDVLRLM K SLTIQRNYVC QGDDGLMIID GTTAGKVNSE TIQKMLELIS KYGEEFGWKY DIAYDGTAEY LKLYFIFGCR IPNLSRHPI VGKERANSSA EEPWPAILDQ IMGVFFNGVH DGLQWQRWIR YSWALCCAFS RQRTMIGESV GYLQYPMWSF VYWGLPLVKA FGSDPWIFS WYMPTGDLGM YSWISLIRPL MTRWMVANGY VTDRCSPVFG NADYRRCFNE LKLYQGYYMA QLPRNPKKSG R AAPREVRE QFTQALSDYL MQNPELKSRV LRGRSEWEKY GAGIIHNPPS LFDVPHKWYQ GAQEAAIATR EELAEMDETL MR ARRHSYS SFSKLLEAYL LVKWRMCEAR EPSVDLRLPL CAGIDPLNSD PFLKMVSVGP MLQSTRKYFA QTLFMAKTVS GLD VNAIDS ALLRLRTLGA DKKALTAQLL MVGLQESEAD ALAGKIMLQD VNTVQLARVV NLAVPDTWMS LDFDSMFKHH VKLL PKDGR HLNTDIPPRM GWLRAILRFL GAGMVMTATG VAVDIYLEDI HGGGRSLGQR FMTWMRQEGR SA |
-Macromolecule #4: Lambda 1
| Macromolecule | Name: Lambda 1 / type: protein_or_peptide / ID: 4 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mammalian orthoreovirus 3 Mammalian orthoreovirus 3 |
| Molecular weight | Theoretical: 122.842859 KDa |
| Sequence | String: YQCHVCSAVL FSPLDLDAHV ASHGLHGNMT LTSSDIQRHI TEFISSWQNH PIVQVSADVE NKKTAQLLHA DTPRLVTWDA GLCTSFKIV PIVPAQVPQD VLAYTFFTSS YAIQSPFPEA AVSRIVVHTR WASNVDFDRD SSVIMAPPTE NNIHLFKQLL N TETLSVRG ...String: YQCHVCSAVL FSPLDLDAHV ASHGLHGNMT LTSSDIQRHI TEFISSWQNH PIVQVSADVE NKKTAQLLHA DTPRLVTWDA GLCTSFKIV PIVPAQVPQD VLAYTFFTSS YAIQSPFPEA AVSRIVVHTR WASNVDFDRD SSVIMAPPTE NNIHLFKQLL N TETLSVRG ANPLMFRANV LHMLLEFVLD NLYLNRHTGF SQDHTPFTEG ANLRSLPGPD AEKWYSIMYP TRMGTPNVSK IC NFVASCV RNRVGRFDRA QMMNGAMSEW VDVFETSDAL TVSIRGRWMA RLARMNINPT EIEWALTECA QGYVTVTSPY APS VNRLMP YRISNAERQI SQIIRIMNIG NNATVIQPVL QDISVLLQRI SPLQIDPTII SNTMSTVSES TTQTLSPASS ILGK LRPSN SDFSSFRVAL AGWLYNGVVT TVIDDSSYPK DGGSVTSLEN LWDFFILALA LPLTTDPCAP VKAFMTLANM MVGFE TIPM DNQIYTQSRR ASAFSTPHTW PRCFMNIQLI SPIDAPILRQ WAEIIHRYWP NPSQIRYGAP NVFGSANLFT PPEVLL LPI DHQPANVTTP TLDFTNELTN WRARVCELMK NLVDNQRYQP GWTQSLVSSM RGTLDKLKLI KSMTPMYLQQ LAPVELA VI APMLPFPPFQ VPYVRLDRDR VPTMVGVTRQ SRDTITQPAL SLSTTNTTVG VPLALDARAI TVALLSGKYP PDLVTNVW Y ADAIYPMYAD TEVFSNLQRD MITCEAVQTL VTLVAQISET QYPVDRYLDW IPSLRASAAT AATFAEWVNT SMKTAFDLS DMLLEPLLSG DPRMTQLAIQ YQQYNGRTFN IIPEMPGSVI ADCVQLTAEV FNHEYNLFGI ARGDIIIGRV QSTHLWSPLA PPPDLVFDR DTPGVHIFGR DCRISFGMNG AAPMIRDETG LMVPFEGNWI FPLALWQMNT RYFNQQFDAW IKTGELRIRI E MGAYPYML HYYDPRQYAN AWNLTSAWLE EITPTSIPSV PFMVPISSDH DISSAPAVQY IISTEYNDRS LFCTNSSSPQ TI AGPDKHI PVERYNILTN PDAPPTQIQL PEVVDLYNVV TRYAYETPPI TAVVMGVP |
-Macromolecule #5: Lambda 1
| Macromolecule | Name: Lambda 1 / type: protein_or_peptide / ID: 5 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mammalian orthoreovirus 3 Mammalian orthoreovirus 3 |
| Molecular weight | Theoretical: 19.112762 KDa |
| Sequence | String: MKRIPRKTKG KSSGKGNDST ERADDGSSQL RDKQNNKAGP ATTEPGTSNR EQYKARPGIA SVQRATESAE MPMKNNDEGT PDKKGNTKG DLVNEHSEAK DEADEATKKQ AKDTDKSKAQ VTYSDTGINN ANELSRSGNV DNEGGSNQKP MSTRIAEATS A IVSKHPAR VGLPPTASSG HG |
-Macromolecule #3: transcript RNA
| Macromolecule | Name: transcript RNA / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Mammalian orthoreovirus 3 Dearing Mammalian orthoreovirus 3 Dearing |
| Molecular weight | Theoretical: 1.263842 KDa |
| Sequence | String: AAGC |
-Macromolecule #6: RNA (60-MER)
| Macromolecule | Name: RNA (60-MER) / type: rna / ID: 6 / Number of copies: 8 |
|---|---|
| Source (natural) | Organism:   Mammalian orthoreovirus 3 Dearing Mammalian orthoreovirus 3 Dearing |
| Molecular weight | Theoretical: 19.016188 KDa |
| Sequence | String: AUAUAUAUAU AUAUAUAUAU AUAUAUAUAU AUAUAUAUAU AUAUAUAUAU AUAUAUAUAU |
-Macromolecule #7: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 56.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller